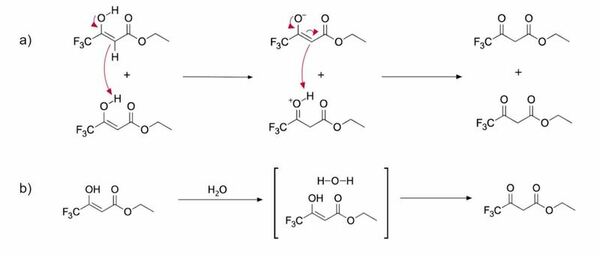
In this study, the authors determined whether tautomerization dynamics in protic and aprotic solvents displayed differences in reaction rates and in the proportion of the keto and enol tautomers present.
Read More...Deuterated solvent effects in the kinetics and thermodynamics of keto-enol tautomerization of ETFAA

In this study, the authors determined whether tautomerization dynamics in protic and aprotic solvents displayed differences in reaction rates and in the proportion of the keto and enol tautomers present.
Read More...Validating DTAPs with large language models: A novel approach to drug repurposing

Here, the authors investigated the integration of large language models (LLMs) with drug target affinity predictors (DTAPs) to improve drug repurposing, demonstrating a significant increase in prediction accuracy, particularly with GPT-4, for psychotropic drugs and the sigma-1 receptor. This novel approach offers to potentially accelerate and reduce the cost of drug discovery by efficiently identifying new therapeutic uses for existing drugs.
Read More...Applying centrality analysis on a protein interaction network to predict colorectal cancer driver genes
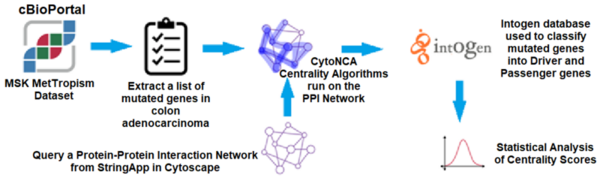
In this article the authors created an interaction map of proteins involved in colorectal cancer to look for driver vs. non-driver genes. That is they wanted to see if they could determine what genes are more likely to drive the development and progression in colorectal cancer and which are present in altered states but not necessarily driving disease progression.
Read More...A novel approach to determine which organism best displays Gijswijt's Sequence in its genome
The sequence of nitrogenous bases that make up the DNA of organisms can contain hidden mathematical sequences. Here the authors used BioPython, a programming tool, to find an organism that displays Gijswijt’s Sequence in its genome. In this manner they found that the common carp best displays Gijswijt’s Sequence in its genome.
Read More...Assessing the accuracy and efficiency of simplified gridded ion thruster simulations
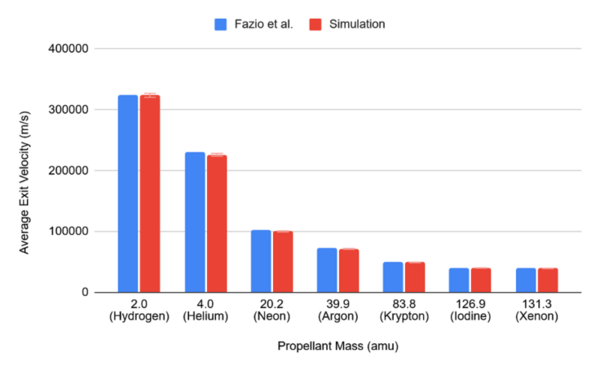
The authors used a particle-in-cell simulation to determine the effects on extensive and intensive metrics. They found that preliminary simulations could be run quickly with much lower particle counts before more technically demanding and comprehensive simulations are performed.
Read More...A comparative analysis of machine learning approaches to predict brain tumors using MRI

The authors use machine learning on MRI images of brain tissue to predict tumor onset as an avenue for early detection of brain cancer.
Read More...Temporal characterization of electroencephalogram slowing activity types
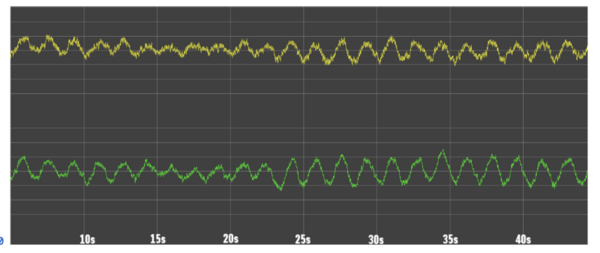
The authors use machine learning to analyze electroencephalogram data and identify slowing patterns that can indicate undetected disorders like epilepsy or dementia
Read More...Convolutional neural network-based analysis of pediatric chest X-ray images for pneumonia detection
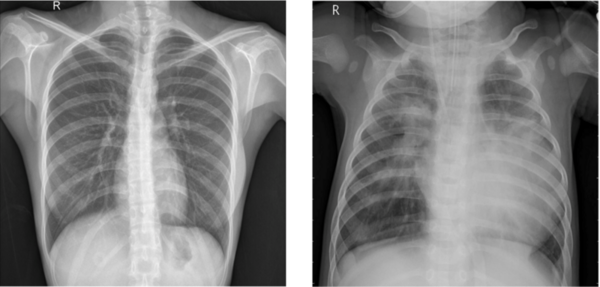
The authors test various machine learning models to improve the accuracy and efficiency of pneumonia diagnosis from X-ray images.
Read More...Quantitative NMR spectroscopy reveals solvent effects in the photochemical degradation of thymoquinone
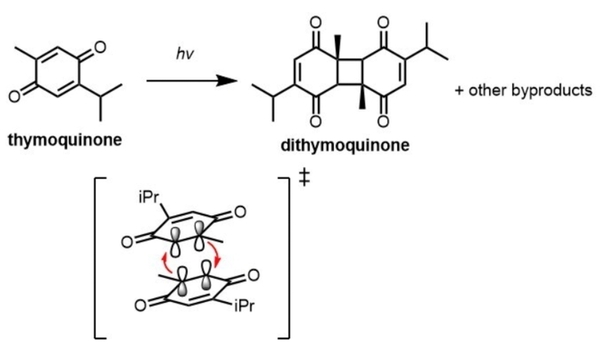
Thymoquinone is a compound of great therapeutic potential and scientific interest. However, its clinical administration and synthetic modifications are greatly limited by its instability in the presence of light. This study employed quantitative 1H nuclear magnetic resonance (NMR) spectroscopy to identify the effect of solvation on the degradation of thymoquinone under ultraviolet light (UV). It found that the rate of degradation is highly solvent dependent occurs maximally in chloroform.
Read More...Hammett linear free-energy relationships in the biocatalytic hydrolysis of para-substituted nitrophenyl benzoate esters
As the world moves towards more eco-friendly methods for chemical synthesis, there's a strong interest in employing enzymes in chemical synthetic processes. Here, the authors explore how the activity of enzymes such as trypsin, lipase and nattokinase is affected by the electronic effects of the substrate they are acting on.
Read More...