Applying centrality analysis on a protein interaction network to predict colorectal cancer driver genes
(1) Stephen F. Austin High School, (2) Department of Neurology, University of California - San Francisco
https://doi.org/10.59720/23-025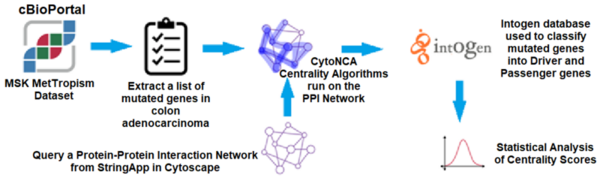
The colorectal cancer tumor microenvironment presents significant genetic heterogeneity with mutations in genes in several signaling pathways. Detecting these driver genes through wet lab experiments is costly and time-consuming. Computational models and bioinformatic tools have become a vital alternative in this effort. One of these novel computational methods, Centrality Analysis, models molecular functions, biological processes and biochemical pathways by creating and analyzing protein-protein interaction networks. Centrality Analysis is an approach to quantify node (in this case, protein) importance in these networks. Essential proteins play critical roles in cell function; therefore, centrality measures serve as a basis to study the relationship between lethality and essentiality by evaluating the topological features of the network. However, there is no established standard to determine the most appropriate centrality measure for analyzing a specific network. The choice of a suitable set is complicated by the impact of network topology because results vary based on network structure, correlation among the selected set of measures, and network data collection methods used. We hypothesized that centrality scores can be used to predict driver genes while statistical and machine learning analyses can identify the relevant centrality features for this task. We proposed different analyses to select a valid set of centrality algorithms to predict driver and non-driver genes. We first recreated a protein-protein interaction network for colorectal cancer featuring known driver and passenger genes, and then compared the centrality scores of eight different algorithms using statistical analysis. We further validated the results by implementing machine learning models. Both analyses identified betweenness and closeness centrality algorithms as most important to predict driver versus non-driver genes.
This article has been tagged with: