
The authors describe the design and testing of new guide RNAs targeting the DMPK gene, which is responsible for myotonic dystrophy.
Read More...Designing gRNAs to reduce the expression of the DMPK gene in patients with classic myotonic dystrophy

The authors describe the design and testing of new guide RNAs targeting the DMPK gene, which is responsible for myotonic dystrophy.
Read More...Identification of potential therapeutic targets for multiple myeloma by gene expression analysis
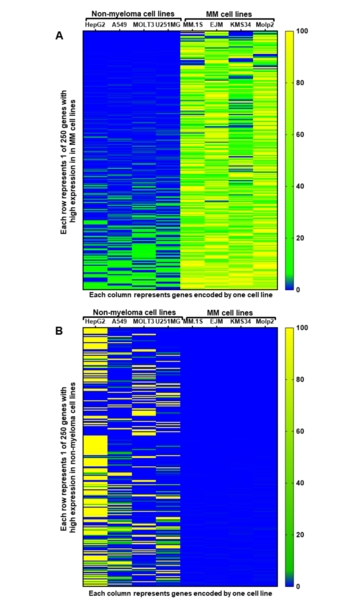
A central challenge of cancer therapy is identifying treatments that will effectively target cancer cells while minimizing effects on healthy cells. To identify potential targets for treating a multiple myeloma, a frequently incurable cancer, Kochenderfer and Kochenderfer analyze RNA sequencing data from the Cancer Cell Line Encyclopedia to find genes with high expression in multiple myeloma cells and low expression in normal tissues
Read More...The effects of the cancer metastasis promoting gene CD151 in E. coli
The independent effects of metastasis-promoting gene CD151 in the process of metastasis are not known. This study aimed to isolate CD151 to discover what its role in metastasis would be uninfluenced by potential interactions with other components and pathways in human cells. Results showed that CD151 significantly increased the adhesion of the cells and decreased their motility. Thus, it may be that CD151 is upregulated in cancer cells for the last step of metastasis, and it increases the chances of success of metastasis by aiding in implantation of the cancer cells. Targeting CD151 in chemotherapeutic modalities could therefore potentially slow or prevent metastasis.
Read More...The Effect of the Human MeCP2 gene on Drosophila melanogaster behavior and p53 inhibition as a model for Rett Syndrome

In this study, the authors observe if the symptoms of Rett Syndrome, a neurodegenerative disease in humans, are reflected in Drosophila melanogaster. This was achieved by differentiating the behavior and physical aspects of wild-type flies from flies expressing the full-length MeCP2 gene and the mutated MeCP2 gene (R106W). After conducting these experiments, some of the Rett Syndrome symptoms were recapitulated in Drosophila, and a subset of those were partially ameliorated by the introduction of pifithrin-alpha.
Read More...String analysis of exon 10 of the CFTR gene and the use of Bioinformatics in determination of the most accurate DNA indicator for CF prediction
Cystic fibrosis is a genetic disease caused by mutations in the CFTR gene. In this paper, the authors attempt to identify variations in stretches of up to 8 nucleotides in the protein-coding portions of the CFTR gene that are associated with disease development. This would allow screening of newborns or even fetuses in utero to determine the likelihood they develop cystic fibrosis.
Read More...Characterization and Phylogenetic Analysis of the Cytochrome B Gene (cytb) in Salvelinus fontinalis, Salmo trutta and Salvelinus fontinalis X Salmo trutta Within the Lake Champlain Basin

Recent declines in the brook trout population of the Lake Champlain Basin have made the genetic screening of this and other trout species of utmost importance. In this study, the authors collected and analyzed 21 DNA samples from Lake Champlain Basin trout populations and performed a phylogenetic analysis on these samples using the cytochrome b gene. The findings presented in this study may influence future habitat decisions in this region.
Read More...Investigating the effects of mutations of amino acids on the protein expression of CDK2 cancer gene
Using CRISPR technology to inhibit the replication of human cytomegalovirus by deletion of a gene promoter
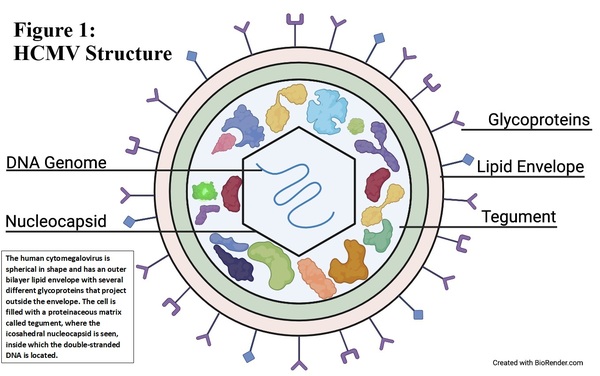
Human cytomegalovirus (HCMV) causes serious infections in immunocompromised patients and therapies to inhibit latent HCMV are not developed. Using CRISPR/Cas9, the authors were able to delete an important promoter region in HCMV.
Read More...Effects on Learning and Memory of a Mutation in Dα7: A D. melanogaster Homolog of Alzheimer's Related Gene for nAChR α7

Alzheimer's disease (AD) involves the reduction of cholinergic activity due to a decrease in neuronal levels of nAChR α7. In this work, Sanyal and Cuellar-Ortiz explore the role of the nAChR α7 in learning and memory retention, using Drosophila melanogaster as a model organism. The performance of mutant flies (PΔEY6) was analyzed in locomotive and olfactory-memory retention tests in comparison to wild type (WT) flies and an Alzheimer's disease model Arc-42 (Aβ-42). Their results suggest that the lack of the D. melanogaster-nAChR causes learning, memory, and locomotion impairments, similar to those observed in Alzheimer's models Arc-42.
Read More...Activated NF-κB Pathway in an Irf6-Deficient Mouse Model for Van der Woude Syndrome
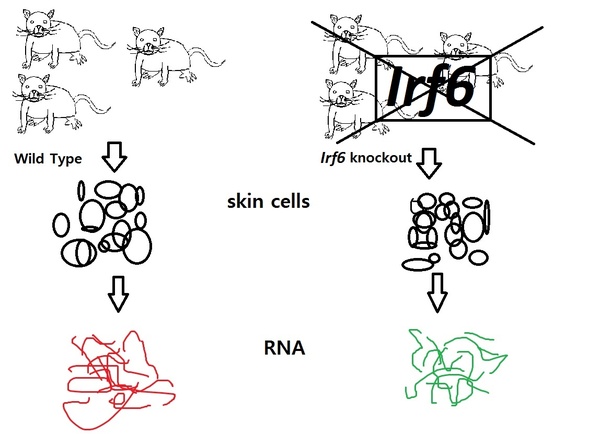
Van der Woude syndrome is a common birth defect caused by mutations in the gene Irf6. In this project, students used microarray expression analysis from wild-type and Irf6-deficient mice in order to identify gene networks or pathways differentially regulated due to the Irf6 mutation. They found NF-κB pathway to be activated in deficient mice.
Read More...