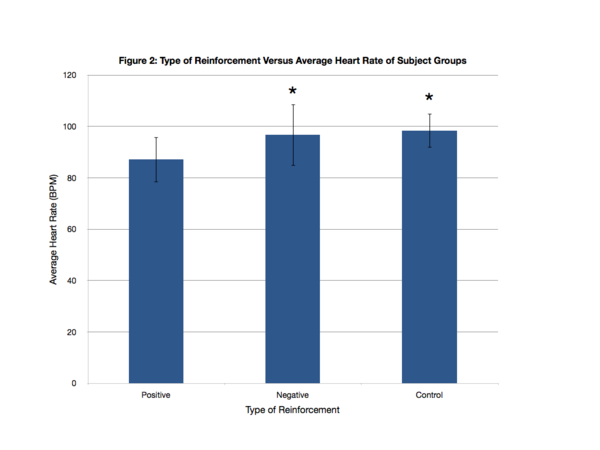
What type of motivation is more effective: reward or punishment? In this study, the authors assess the effects of positive or negative on the math scores of sixth graders.
Read More...The Effect of Positive and Negative Reinforcement on Sixth Graders’ Mental Math Performance

What type of motivation is more effective: reward or punishment? In this study, the authors assess the effects of positive or negative on the math scores of sixth graders.
Read More...Recognition of animal body parts via supervised learning
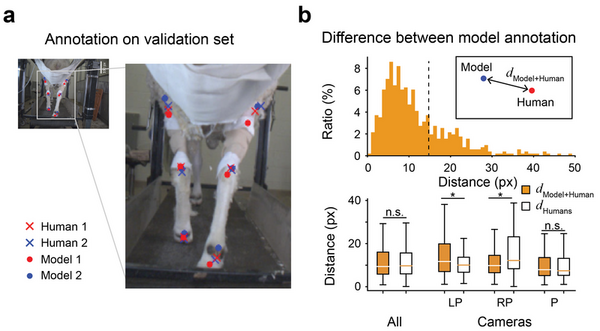
The application of machine learning techniques has facilitated the automatic annotation of behavior in video sequences, offering a promising approach for ethological studies by reducing the manual effort required for annotating each video frame. Nevertheless, before solely relying on machine-generated annotations, it is essential to evaluate the accuracy of these annotations to ensure their reliability and applicability. While it is conventionally accepted that there cannot be a perfect annotation, the degree of error associated with machine-generated annotations should be commensurate with the error between different human annotators. We hypothesized that machine learning supervised with adequate human annotations would be able to accurately predict body parts from video sequences. Here, we conducted a comparative analysis of the quality of annotations generated by humans and machines for the body parts of sheep during treadmill walking. For human annotation, two annotators manually labeled six body parts of sheep in 300 frames. To generate machine annotations, we employed the state-of-the-art pose-estimating library, DeepLabCut, which was trained using the frames annotated by human annotators. As expected, the human annotations demonstrated high consistency between annotators. Notably, the machine learning algorithm also generated accurate predictions, with errors comparable to those between humans. We also observed that abnormal annotations with a high error could be revised by introducing Kalman Filtering, which interpolates the trajectory of body parts over the time series, enhancing robustness. Our results suggest that conventional transfer learning methods can generate behavior annotations as accurate as those made by humans, presenting great potential for further research.
Read More...Refinement of Single Nucleotide Polymorphisms of Atopic Dermatitis related Filaggrin through R packages

In the United States, there are currently 17.8 million affected by atopic dermatitis (AD), commonly known as eczema. It is characterized by itching and skin inflammation. AD patients are at higher risk for infections, depression, cancer, and suicide. Genetics, environment, and stress are some of the causes of the disease. With the rise of personalized medicine and the acceptance of gene-editing technologies, AD-related variations need to be identified for treatment. Genome-wide association studies (GWAS) have associated the Filaggrin (FLG) gene with AD but have not identified specific problematic single nucleotide polymorphisms (SNPs). This research aimed to refine known SNPs of FLG for gene editing technologies to establish a causal link between specific SNPs and the diseases and to target the polymorphisms. The research utilized R and its Bioconductor packages to refine data from the National Center for Biotechnology Information's (NCBI's) Variation Viewer. The algorithm filtered the dataset by coding regions and conserved domains. The algorithm also removed synonymous variations and treated non-synonymous, frameshift, and nonsense separately. The non-synonymous variations were refined and ordered by the BLOSUM62 substitution matrix. Overall, the analysis removed 96.65% of data, which was redundant or not the focus of the research and ordered the remaining relevant data by impact. The code for the project can also be repurposed as a tool for other diseases. The research can help solve GWAS's imprecise identification challenge. This research is the first step in providing the refined databases required for gene-editing treatment.
Read More...Blockchain databases: Encrypted for efficient and secure NoSQL key-store
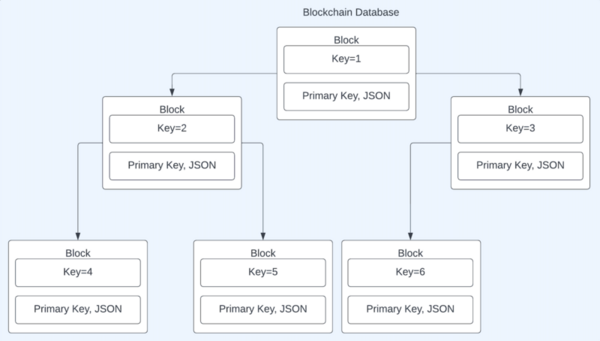
Although commonly associated with cryptocurrency, blockchains offer security that other databases could benefit from. These student authors tested a blockchain database framework, and by tracking runtime of four independent variables, they prove this framework is feasible for application.
Read More...Hammett linear free-energy relationships in the biocatalytic hydrolysis of para-substituted nitrophenyl benzoate esters
As the world moves towards more eco-friendly methods for chemical synthesis, there's a strong interest in employing enzymes in chemical synthetic processes. Here, the authors explore how the activity of enzymes such as trypsin, lipase and nattokinase is affected by the electronic effects of the substrate they are acting on.
Read More...In silico modeling of emodin’s interactions with serine/threonine kinases and chitosan derivatives

Here, through protein-ligand docking, the authors investigated the effect of the interaction of emodin with serine/threonine kinases, a subclass of kinases that is overexpressed in many cancers, which is implicated in phosphorylation cascades. Through molecular dynamics theyfound that emodin forms favorable interactions with chitosan and chitosan PEG (polyethylene glycol) copolymers, which could aid in loading drugs into nanoparticles (NPs) for targeted delivery to cancerous tissue. Both polymers demonstrated reasonable entrapment efficiencies, which encourages experimental exploration of emodin through targeted drug delivery vehicles and their anticancer activity.
Read More...Effect of Increasing Concentrations of Cannabidiol (CBD) on Hatching, Survival and Development of Artemia salina

Cannabidiol, or CBD, is a widely available over the counter treatment used for various medical conditions. However, CBD exerts its effects on the endocannabinoid system, which is involved in neural maturation, and could potentially have adverse effects on brain development. Here, the impact of CBD on the development of brine shrimp (Artemia salina) was assessed. Differences in dose responses were observed.
Read More...Comparing Virulence of Three T4 Bacteriophage Strains on Ampicillin-Resistant and Sensitive E. coli Bacteria
In this study, the authors investigate an alternative way to kill bacteria other than the use of antibiotics, which is useful when considering antibiotic-resistance bacteria. They use bacteriophages, which are are viruses that can infect bacteria, and measure cell lysis. They make some important findings that these bacteriophage can lyse both antibiotic-resistant and non-resistant bacteria.
Read More...Impact of NaCl concentration in crystalline nanocellulose for printed ionic dielectrics
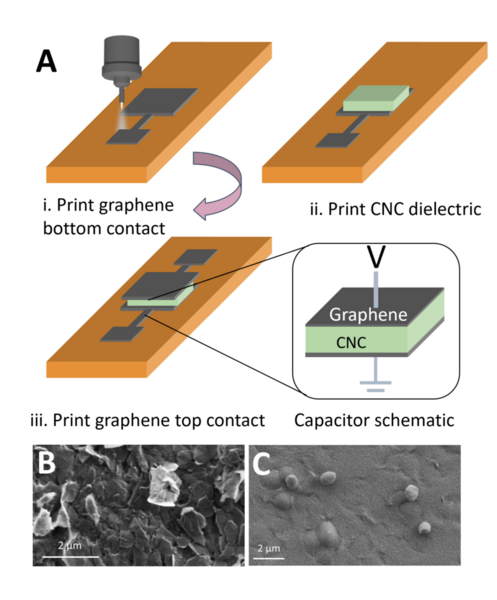
The authors looked at how the addition of NaCl to crystalline nanocellulose capacitors could improve performance in transistor applications. They found that NaCl can improve performance, but that further work is needed to determine the optimal concentration used depending on the intended application.
Read More...Impact of aluminum surface area on the rate of reaction with aqueous copper (II) chloride solutions

In this article the authors looked at how temperature was impacted when alumnium was added in various forms to aqueous copper(II) solutions. Their study investigates the impact of surface area on chemical reactions.
Read More...Search articles by title, author name, or tags