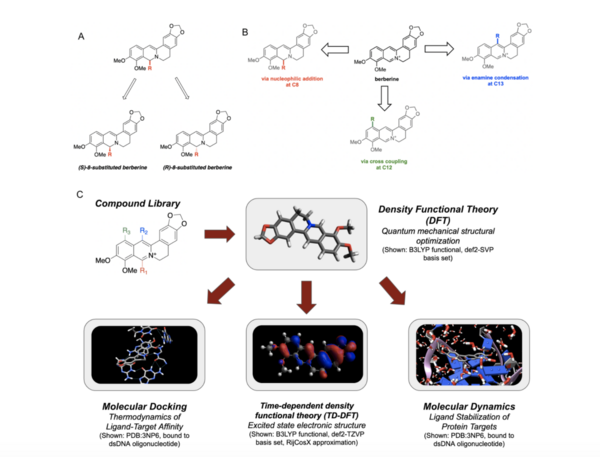
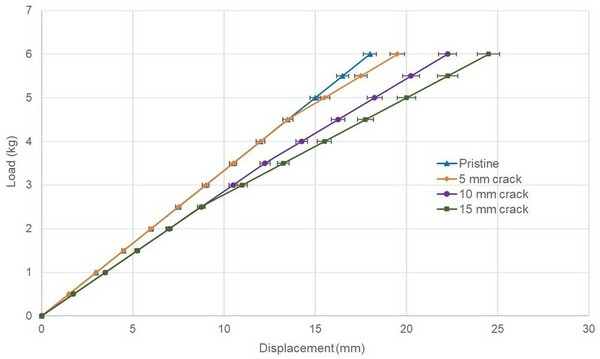
Berberine, a natural product alkaloid, and its analogs have a wide range of medicinal properties, including antibacterial and anticancer effects. Here, the authors explored a library of alkyl or aryl berberine analogs to probe binding to double-stranded and G-quadruplex DNA. They determined that the nature of the substituent, the position of the substituent, and the nucleic acid target affect the free energy of binding of berberine analogs to DNA and G-quadruplex DNA, however berberine analogs did not result in net stabilization of G-quadruplex DNA.
Read More...
.png)