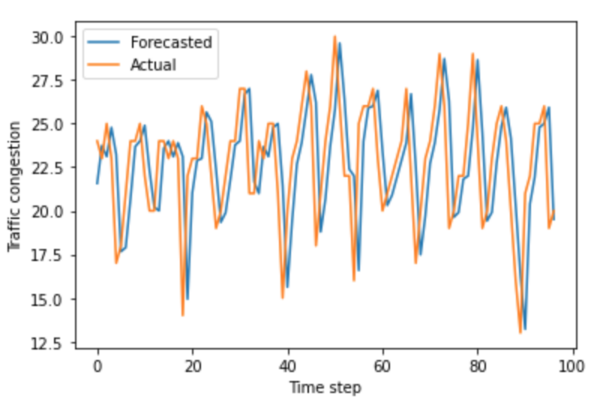
In this paper, we measured the privacy budgets and utilities of different differentially private mechanisms combined with different machine learning models that forecast traffic congestion at future timestamps. We expected the ANNs combined with the Staircase mechanism to perform the best with every value in the privacy budget range, especially with the medium high values of the privacy budget. In this study, we used the Autoregressive Integrated Moving Average (ARIMA) and neural network models to forecast and then added differentially private Laplacian, Gaussian, and Staircase noise to our datasets. We tested two real traffic congestion datasets, experimented with the different models, and examined their utility for different privacy budgets. We found that a favorable combination for this application was neural networks with the Staircase mechanism. Our findings identify the optimal models when dealing with tricky time series forecasting and can be used in non-traffic applications like disease tracking and population growth.
Read More...