
The authors describe the design and testing of new guide RNAs targeting the DMPK gene, which is responsible for myotonic dystrophy.
Read More...Designing gRNAs to reduce the expression of the DMPK gene in patients with classic myotonic dystrophy

The authors describe the design and testing of new guide RNAs targeting the DMPK gene, which is responsible for myotonic dystrophy.
Read More...Design and in silico screening of analogs of rilpivirine as novel non-nucleoside reverse transcriptase inhibitors (NNRTIs) for antiretroviral therapy
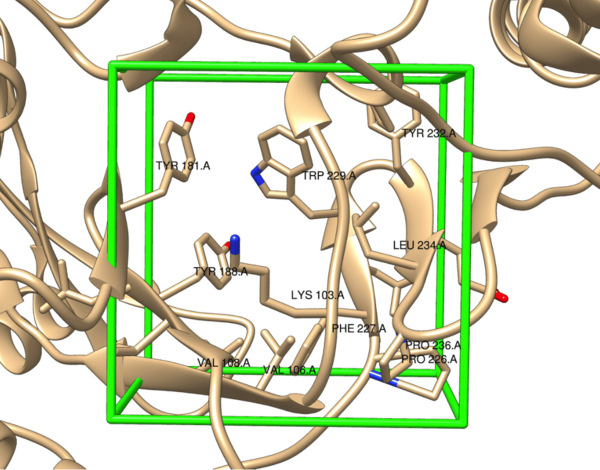
In this study, the authors use high-throughput virtual screening to design and evaluate a set of non-nucleoside reverse transcriptase inhibitors for binding affinity to the protein reverse transcriptase. These studies have important applications toward HIV therapies.
Read More...Extracellular vesicles derived from oxidatively stressed stromal cells promote cancer progression
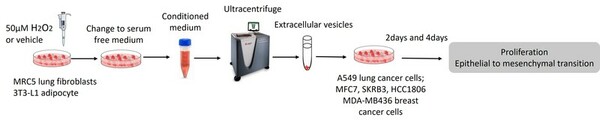
This paper hypothesized that the tumor microenvironment mediates cancer’s response to oxidative stress by delivering extracellular vesicles to cancer cells. Breast and lung cancer cells were treated with EVs, reavealing that EVs extracted from oxidatively stressed adipocytes increased the cell proliferation of breast cancer cells. These findings present a novel way that the TME influences cancer progression.
Read More...Innovative Treatment for Reducing Senescence and Revitalizing Aging Cells through Gene Silencing
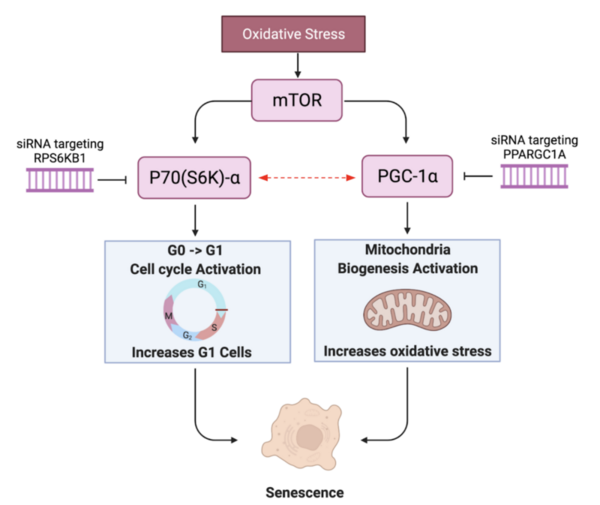
Cellular senescence plays a key role in aging cells and is attributed to a number of disease and pathology. These authors find that genetic editing of both RPS6KB1 and PPARGC1A revitalizes a human skin fibroblast cell line.
Read More...Application of gene therapy for reversing T-cell dysfunction in cancer
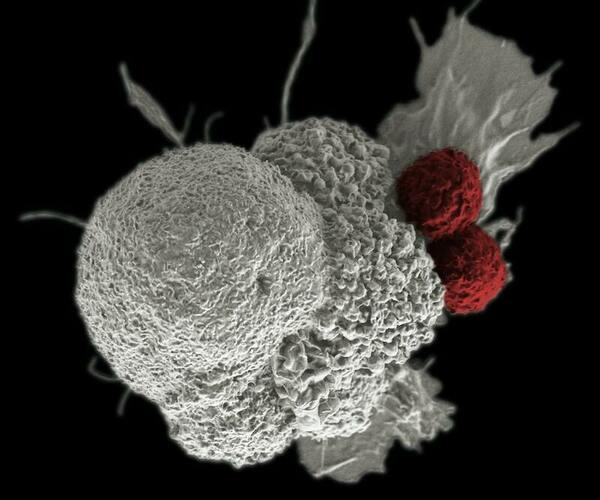
Since cancer cells inhibit T-cell activity, the authors investigated a method to reverse T-cell disfunction with gene therapy, so that the T-cells would become effective once again in fighting cancer cells. They used the inhibition of proprotein convertases (PCSK1) in T cells and programmed death-ligand 1 (CD274) in cancer cells. They observed the recovery of IL-2 expression in Jurkat cells, with increased recovery noted in a co-culture sample. This study suggests a novel strategy to reactivate T cells.
Read More...RNAi-based Gene Therapy Targeting ZGPAT Promotes EGF-dependent Wound Healing
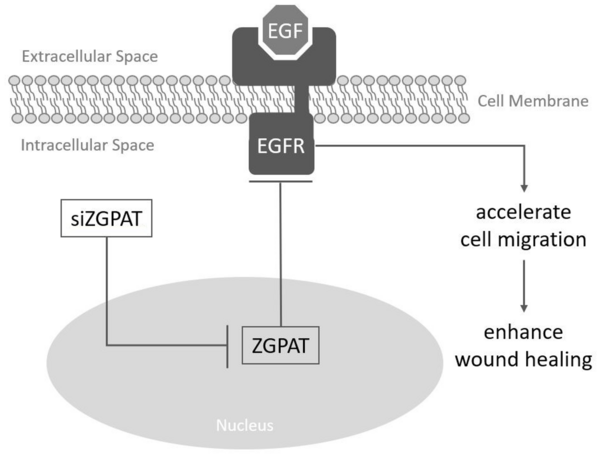
Wound-healing involves a sequence of events, such as inflammation, proliferation, and migration of different cell types like fibroblasts. Zinc Finger CCCH-type with G-Patch Domain Containing Protein (ZGPAT), encodes a protein that has its main role as a transcription repressor by binding to a specific DNA sequence. The aim of the study was to find out whether inhibiting ZGPAT will expedite the wound healing process by accelerating cell migration. This treatment strategy can provide a key to the development of wound healing strategies in medicine and cellular biology.
Read More...Characterization of Inflammatory Cytokine Gene Expression in a Family with a History of Psoriasis
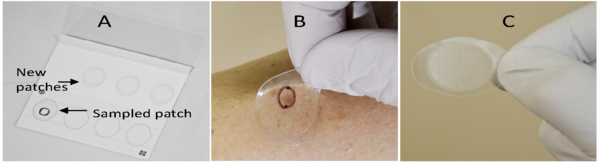
Psoriasis is a heritable autoimmune disorder characterized by abnormal red and itchy skin patches. The authors study the family of a man with psoriasis. They explore whether the man's children, who do not show any symptoms of psoriasis, demonstrate gene expression consistent with the disease.
Read More...Post-Traumatic Stress Disorder (PTSD) biomarker identification using a deep learning model
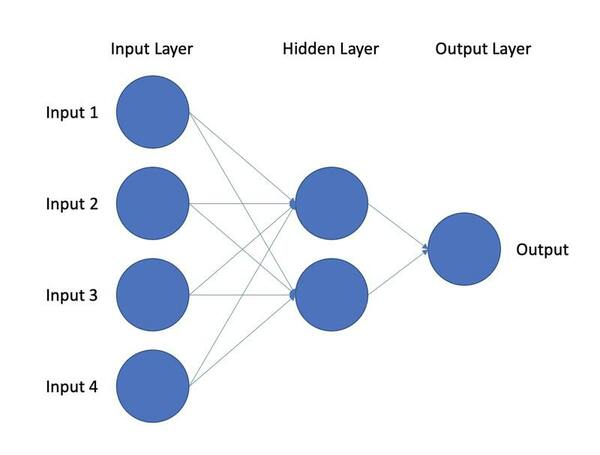
In this study, a deep learning model is used to classify post-traumatic stress disorder patients through novel markers to assist in finding candidate biomarkers for the disorder.
Read More...Gene expression profiling of MERS-CoV-London strain
%20(1).png)
In this study, the authors identify transcripts and gene networks that are changed after infection with the Middle East Respiratory Syndrome-related coronavirus (MERS-CoV).
Read More...