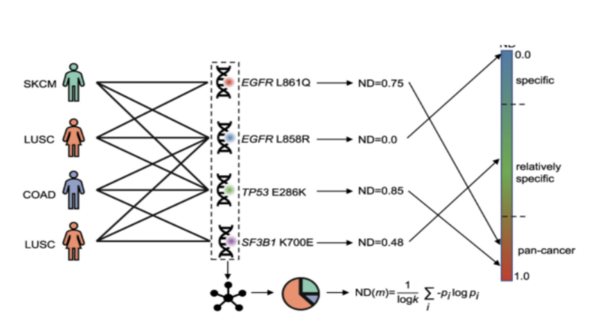
Breast cancer is the most common cancer in women, with approximately 300,000 diagnosed with breast cancer in 2023. It ranks second in cancer-related deaths for women, after lung cancer with nearly 50,000 deaths. Scientists have identified important genetic mutations in genes like BRCA1 and BRCA2 that lead to the development of breast cancer, but previous studies were limited as they focused on specific populations. To overcome limitations, diverse populations and powerful statistical methods like genome-wide association studies and whole-genome sequencing are needed. Explainable artificial intelligence (XAI) can be used in oncology and breast cancer research to overcome these limitations of specificity as it can analyze datasets of diagnosed patients by providing interpretable explanations for identified patterns and predictions. This project aims to achieve technological and medicinal goals by using advanced algorithms to identify breast cancer subtypes for faster diagnoses. Multiple methods were utilized to develop an efficient algorithm. We hypothesized that an XAI approach would be best as it can assign scores to genes, specifically with a 90% success rate. To test that, we ran multiple trials utilizing XAI methods through the identification of class-specific and patient-specific key genes. We found that the study demonstrated a pipeline that combines multiple XAI techniques to identify potential biomarker genes for breast cancer with a 95% success rate.
Read More...