Mismatch repair is not correlated with genomic alterations in glioblastoma patients
(1) Hillsborough High School, (2) Department of Biological Sciences, Indian Institute of Science Education and Research, (3) SkoolMentor.com (Mentorconnect)
https://doi.org/10.59720/22-226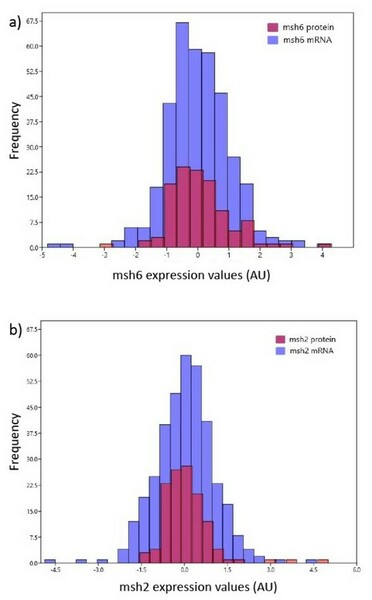
Glioma is the most common brain tumor of the glial cells with a highly infiltrative nature. A grade IV glioblastoma is the most malignant and aggressive presenting with complex mutations and a very high genomic alteration status. Glioblastoma patients have the fastest progression compared to other cancer patients and poor survival. Methylation status of MGMT, the gene encoding a DNA repair enzyme, is the only known promising biomarker. There is a need to look for new biomarkers in similar functional pathways. Two mismatch repair pathway (MMR) genes, hMSH2, and hMSH6, which function downstream of MGMT, appear as ideal targets. We hypothesized that a lower expression of hMSH2 and hMSH6 are associated with a higher fraction of genome-altered (FGA) which would influence poor survival. We conducted an in-silico study using publicly available datasets consisting of clinical and experimental data. We obtained a weak positive correlation between hMSH2 and hMSH6 with FGA. Sex specific analyses yielded a stronger significant positive correlation. However, overall the measures of correlation were too small as the component analysis indicated homology and validated the findings. Additionally, the expression of hMSH2 and hMSH6 did not affect overall survival. We conclude that in this cohort hMSH2 and hMSH6 did not indicate biological significance in causing FGA, nor were they effective biomarkers for survival as hypothesized.
This article has been tagged with: