In this study, the authors analyze gene expression datasets to determine if there is a core set of genes dysregulated during nonalcoholic steatohepatitis.
Read More...Browse Articles
Ribosome distribution affects stalling in amino-acid starved cancer cells
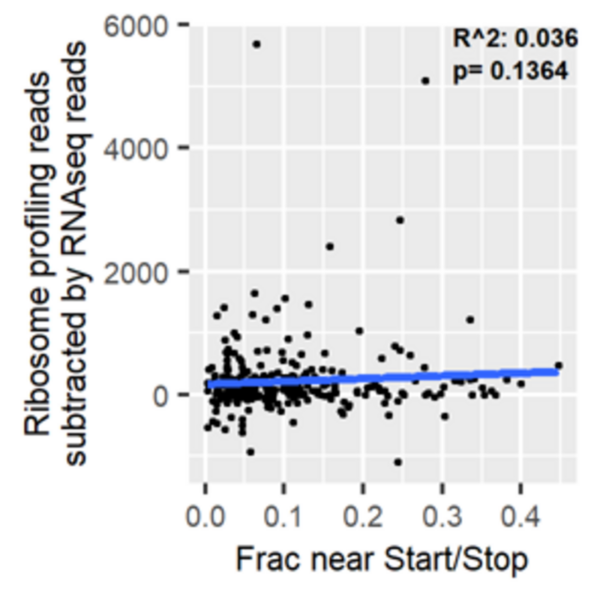
In this article, the authors analyzed ribosome profiling data from amino acid-starved pancreatic cancer cells to explore whether the pattern of ribosome distribution along transcripts under normal conditions can predict the degree of ribosome stalling under stress. The authors found that ribosomes in amino acid-deprived cells stalled more along elongation-limited transcripts. By contrast, they observed no relationship between read density near start and stop and disparities between mRNA sequencing reads and ribosome profiling reads. This research identifies an important relationship between read distribution and propensity for ribosomes to stall, although more work is needed to fully understand the patterns of ribosome distribution along transcripts in ribosome profiling data.
Read More...Contrasting role of ASCC3 and ALKBH3 in determining genomic alterations in Glioblastoma Multiforme
Glioblastoma Multiforme (GBM) is the most malignant brain tumor with the highest fraction of genome alterations (FGA), manifesting poor disease-free status (DFS) and overall survival (OS). We explored The Cancer Genome Atlas (TCGA) and cBioportal public dataset- Firehose legacy GBM to study DNA repair genes Activating Signal Cointegrator 1 Complex Subunit 3 (ASCC3) and Alpha-Ketoglutarate-Dependent Dioxygenase AlkB Homolog 3 (ALKBH3). To test our hypothesis that these genes have correlations with FGA and can better determine prognosis and survival, we sorted the dataset to arrive at 254 patients. Analyzing using RStudio, both ASCC3 and ALKBH3 demonstrated hypomethylation in 82.3% and 61.8% of patients, respectively. Interestingly, low mRNA expression was observed in both these genes. We further conducted correlation tests between both methylation and mRNA expression of these genes with FGA. ASCC3 was found to be negatively correlated, while ALKBH3 was found to be positively correlated, potentially indicating contrasting dysregulation of these two genes. Prognostic analysis showed the following: ASCC3 hypomethylation is significant with DFS and high ASCC3 mRNA expression to be significant with OS, demonstrating ASCC3’s potential as disease prediction marker.
Read More...Reducing PMA-induced COX-2 expression using a herbal formulation in MCF-7 breast cancer cells
In this study, the authors investigate the effect of a herbal formulation on Cyclooxygenase-2 (COX-2) expression in cancer cells. High levels of COX-2 correlates with worsened cancer outcomes and the authors hypothesize that the formulation will inhibit COX-2 levels.
Read More...Disruptions in protein-protein interactions between HTT, PRPF40B, and MECP2 are involved in Lopes-Maciel-Rodan syndrome
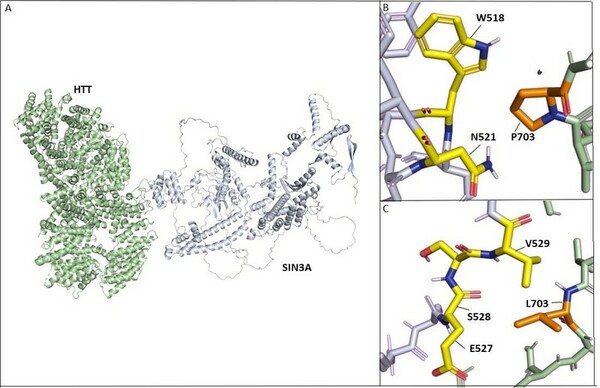
In an extensive study of gene mutations, and their resulting effect on protein-protein interactions, Desai and Stork found that HTT-PRPF40B-MECP2 interactions are weakened with progression of Lopes-Maciel-Rodan syndrome.
Read More...Protective effect of bromelain and pineapple extracts on UV-induced damage in human skin cells

In this study, the authors tested whether the compound bromelain extracted from pineapples could protect skin cells from UV damage.
Read More...