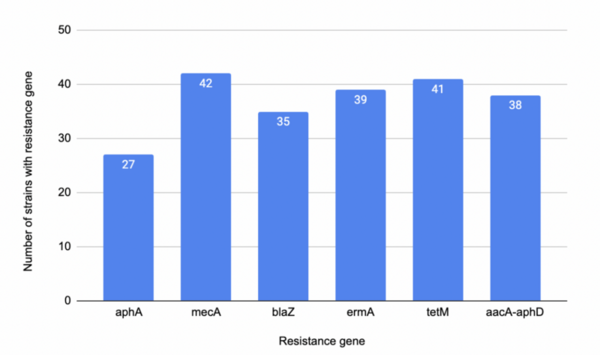
The authors looked at how the presence of difference antibiotic resistance genes would influence antibiotic resistance in Staphylococcus aureus.
Read More...Analysis of antibiotic resistance genes in publicly accessible Staphylococcus aureus genomes

The authors looked at how the presence of difference antibiotic resistance genes would influence antibiotic resistance in Staphylococcus aureus.
Read More...Expression of Anti-Neurodegeneration Genes in Mutant Caenorhabditis elegans Using CRISPR-Cas9 Improves Behavior Associated With Alzheimer’s Disease
Alzheimer's disease is one of the leading causes of death in the United States and is characterized by neurodegeneration. Mishra et al. wanted to understand the role of two transport proteins, LRP1 and AQP4, in the neurodegeneration of Alzheimer's disease. They used a model organism for Alzheimer's disease, the nematode C. elegans, and genetic engineering to look at whether they would see a decrease in neurodegeneration if they increased the amount of these two transport proteins. They found that the best improvements were caused by increased expression of both transport proteins, with smaller improvements when just one of the proteins is overly expressed. Their work has important implications for how we understand neurodegeneration in Alzheimer's disease and what we can do to slow or prevent the progression of the disease.
Read More...Plasmid Variance and Nutrient Regulation of Bioluminescence Genes
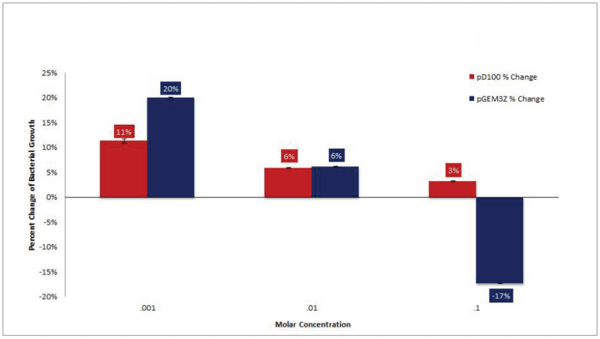
Numerous organisms, including the marine bacterium Aliivibrio fischeri, produce light. This bioluminescence is involved in many important symbioses and may one day be an important source of light for humans. In this study, the authors investigated ways to increase bioluminescence production from the model organism E. coli.
Read More...Expressional correlations between SERPINA6 and pancreatic ductal adenocarcinoma-linked genes
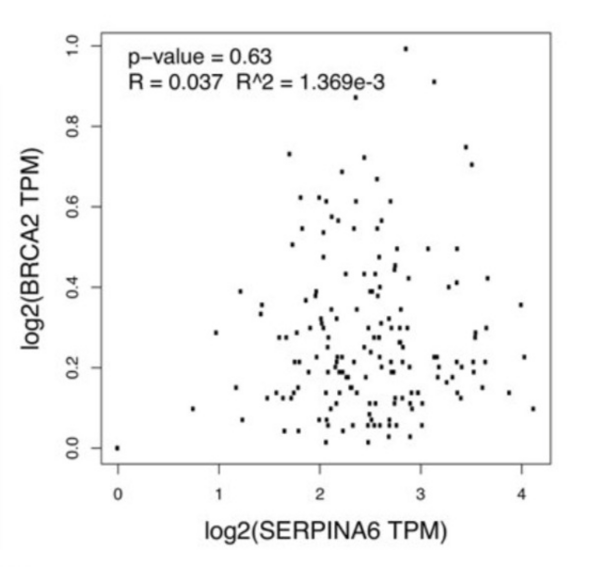
Pancreatic ductal adenocarcinoma (PDAC) is the most common form of pancreatic cancer, with early diagnosis and treatment challenges. When any of the genes KRAS, SMAD4, TP53, and BRCA2 are heavily mutated, they correlate with PDAC progression. Cellular stress, partly regulated by the gene SERPINA6, also correlates with PDAC progression. When SERPINA6 is highly expressed, corticosteroid-binding globulin inhibits the effect of the stress hormone cortisol. In this study, the authors explored whether there is an inverse correlation between the expression of SERPINA6 and PDAC-linked genes.
Read More...Applying centrality analysis on a protein interaction network to predict colorectal cancer driver genes
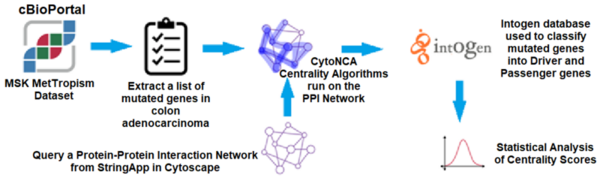
In this article the authors created an interaction map of proteins involved in colorectal cancer to look for driver vs. non-driver genes. That is they wanted to see if they could determine what genes are more likely to drive the development and progression in colorectal cancer and which are present in altered states but not necessarily driving disease progression.
Read More...The effects of dysregulated ion channels and vasoconstriction in glioblastoma multiforme
Gene expression analysis of febrile seizure’s impact on mesial temporal lobe epilepsy
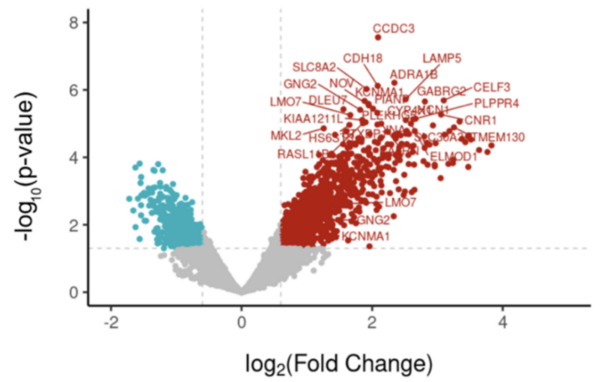
The authors looked at genes that were differentially expressed in patients who had and had not had febrile seizures to determine what differences in gene expression existed.
Read More...DNA repair protein mutations alter blood cancer sensitivity to cisplatin or gemcitabine in vitro
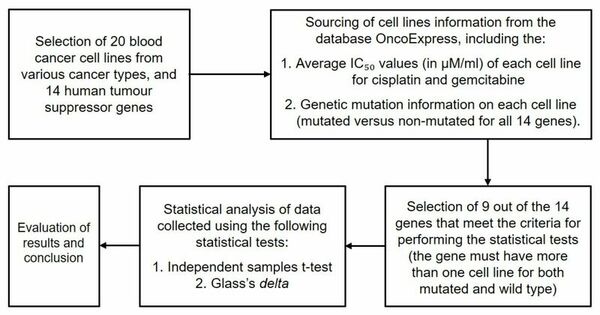
The authors investigate whether human blood cancers carrying mutations in DNA repair genes possess increased sensitivity to common chemotherapy drugs cisplatin or gemcitabine.
Read More...Predicting smoking status based on RNA sequencing data
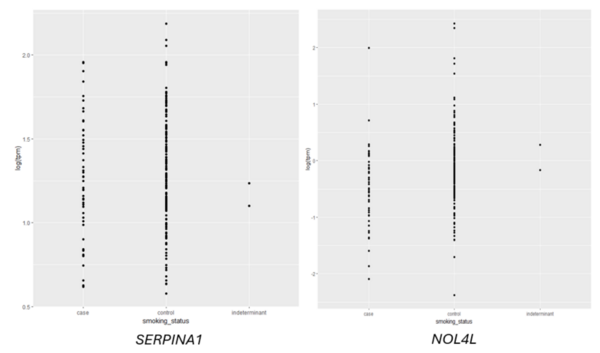
Given an association between nicotine addiction and gene expression, we hypothesized that expression of genes commonly associated with smoking status would have variable expression between smokers and non-smokers. To test whether gene expression varies between smokers and non-smokers, we analyzed two publicly-available datasets that profiled RNA gene expression from brain (nucleus accumbens) and lung tissue taken from patients identified as smokers or non-smokers. We discovered statistically significant differences in expression of dozens of genes between smokers and non-smokers. To test whether gene expression can be used to predict whether a patient is a smoker or non-smoker, we used gene expression as the training data for a logistic regression or random forest classification model. The random forest classifier trained on lung tissue data showed the most robust results, with area under curve (AUC) values consistently between 0.82 and 0.93. Both models trained on nucleus accumbens data had poorer performance, with AUC values consistently between 0.65 and 0.7 when using random forest. These results suggest gene expression can be used to predict smoking status using traditional machine learning models. Additionally, based on our random forest model, we proposed KCNJ3 and TXLNGY as two candidate markers of smoking status. These findings, coupled with other genes identified in this study, present promising avenues for advancing applications related to the genetic foundation of smoking-related characteristics.
Read More...Identifying 5-hydroxymethylcytosine as a potential cancer biomarker using FFPE DNA samples

This study used an improved CMS-seq method to profile 5hmC in ormalin-fixed and paraffin-embedded (FFPE) samples from HNC tumors and adjacent normal tissues, identifying three genes (PRKD2, HADHA, and AIPL1) with promising potential as biomarkers for Head and neck cancer (HNC) diagnosis.
Read More...