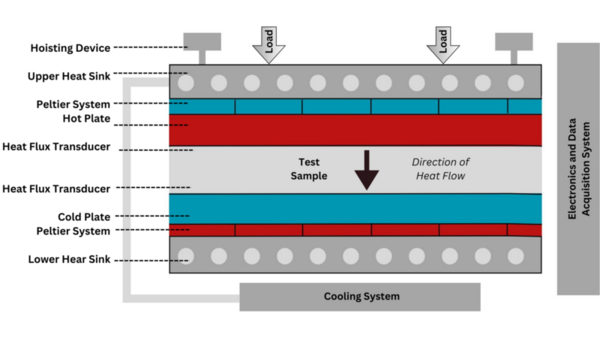
The authors looked into eco-friendly alternatives for insulating material. They ultimately found that a polyurethane derived from eggshells was an effective insulator and further research into it is warranted.
Read More...Investigating sustainable insulation materials: Analysis of biofoams and petroleum-derived foams

The authors looked into eco-friendly alternatives for insulating material. They ultimately found that a polyurethane derived from eggshells was an effective insulator and further research into it is warranted.
Read More...Elevated GPx4 and FSP1 expression in MG63 cells: Exploring potential links to drug resistance and ferroptosis
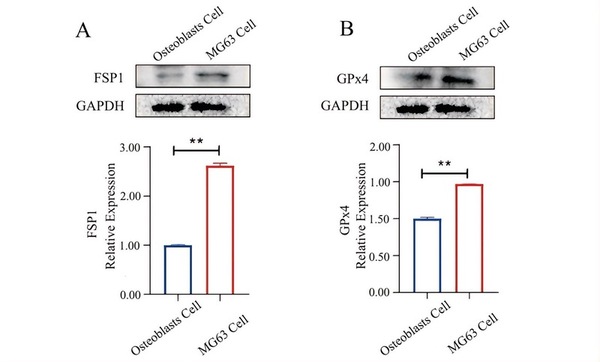
Current osteosarcoma (OS) treatments rely on surgery and chemotherapy, but drug resistance remains a major challenge that lowers patient survival rates. Ferroptosis, a form of regulated cell death, has shown promise in cancer therapy but is not well understood in OS. This study explores the use of Ferroptosis in OS.
Read More...Computational Study of Erosion Effects on a Triangular Aerofoil's Aerodynamics at Reynolds number of 10,000
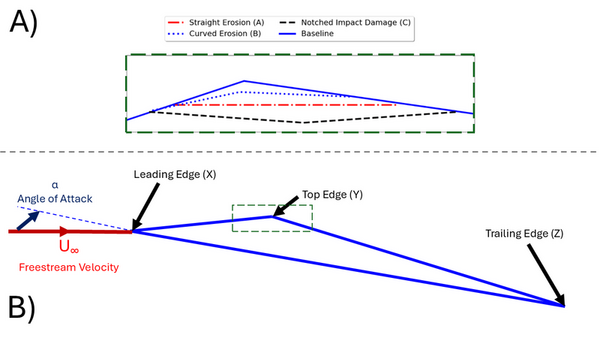
This study examined the impact of erosion on the performance of a triangular aerofoil at a low Reynolds number (Re = 10,000), relevant for harsh conditions like those on Mars.
Read More...Lettuce seed germination in the presence of microplastic contamination

Microplastic pollution is a pressing environmental issue, particularly in the context of its potential impacts on ecosystems and human health. In this study, we explored the ability of plants, specifically those cultivated for human consumption, to absorb microplastics from their growing medium. We found no evidence of microplastic absorption in both intact and mechanically damaged roots. This outcome suggests that microplastics larger than 10 μm may not be readily absorbed by the root systems of leafy crops such as lettuce (L. sativa).
Read More...Investigating Lemna minor and microorganisms for the phytoremediation of nanosilver and microplastics
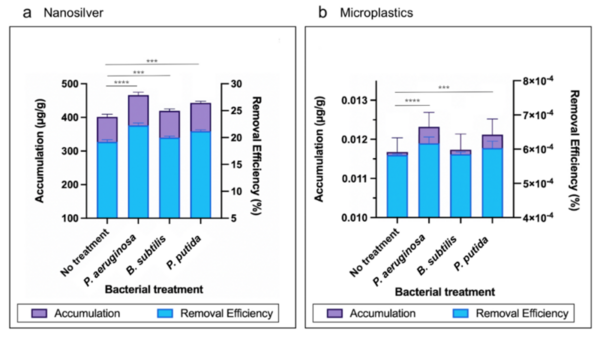
The authors looked at phytoremediation, the process by which plants are used to remove pollutants from our environment, and the ability of Lemna minor to perform phytoremediation in various simulated polluted environments. The authors found that L. minor could remove pollutants from the environment and that the addition of bacteria increased this removal.
Read More...Genetic algorithm based features selection for predicting the unemployment rate of India

The authors looked at using genetic algorithms to look at the Indian labor market and what features might best explain any variation seen. They found that features such as economic growth and household consumption, among others, best explained variation.
Read More...Evaluation of in vitro anti-inflammatory effect of PLAY® on UC-MSCs: A COX-2 expression study
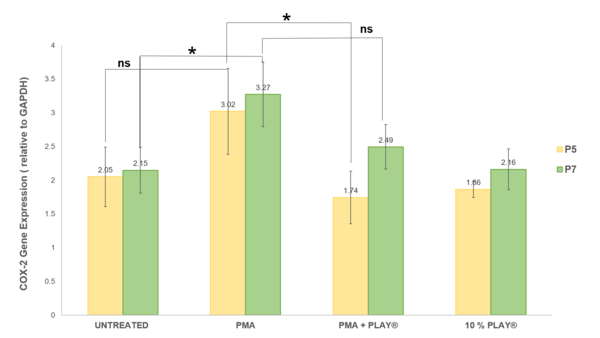
The authors seek to accelerate wound healing by reducing inflammation with a cocktail containing growth factors and bioactive modulators.
Read More...Associations between substance misuse, social factors, depression, and anxiety among college students

Here, the authors considered the effects of relationship status and substance use on the mental health of colleges students, where they specifically examined their correlation with depression, anxiety, and the fear of missing out (FoMO). Through a survey of college students they found that those with higher substance misuse had higher levels of anxiety, depression, and FoMO, while those involved in longer-term relationships had lower levels of FoMo and alcohol use.
Read More...Effects of copper sulfate exposure on the nervous system of the Hirudo verbana leech
In this study, the authors test whether excess copper exposure has neurobehavioral effects on Hirudo verbana leeches.
Read More...Determining surface tension of various liquids and shear modulus of paper using crumpling effect
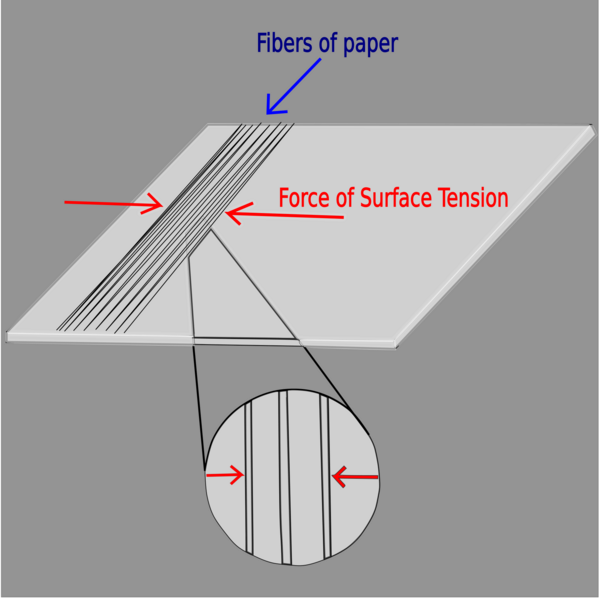
In this article, the authors investigate the shear modulus of different types of paper in the setting of the crumpling effect.
Read More...