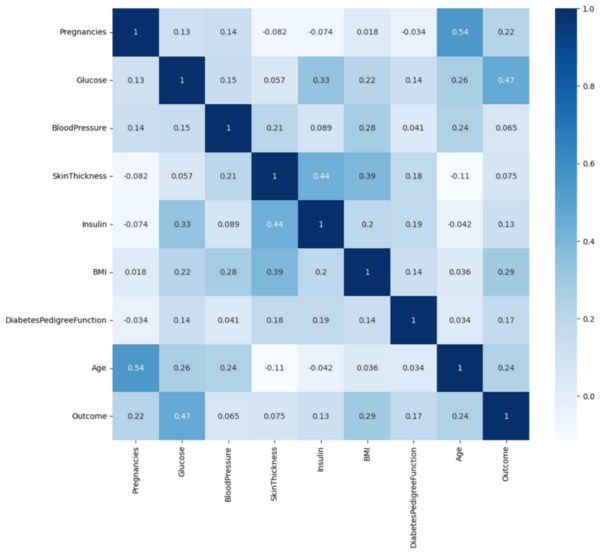
The authors develop and test a machine learning algorithm for predicting diabetes diagnoses.
Read More...Prediction of diabetes using supervised classification

The authors develop and test a machine learning algorithm for predicting diabetes diagnoses.
Read More...A machine learning approach to detect renal calculi by studying the physical characteristics of urine
The authors trained a machine learning model to detect kidney stones based on characteristics of urine. This method would allow for detection of kidney stones prior to the onset of noticeable symptoms by the patient.
Read More...Pruning replay buffer for efficient training of deep reinforcement learning

Reinforcement learning (RL) is a form of machine learning that can be harnessed to develop artificial intelligence by exposing the intelligence to multiple generations of data. The study demonstrates how reply buffer reward mechanics can inform the creation of new pruning methods to improve RL efficiency.
Read More...Artificial intelligence assisted violin performance learning

In this study the authors looked at the ability of artificial intelligence to detect tempo, rhythm, and intonation of a piece played on violin. Technology such as this would allow for students to practice and get feedback without the need of a teacher.
Read More...Statistical models for identifying missing and unclear signs of the Indus script
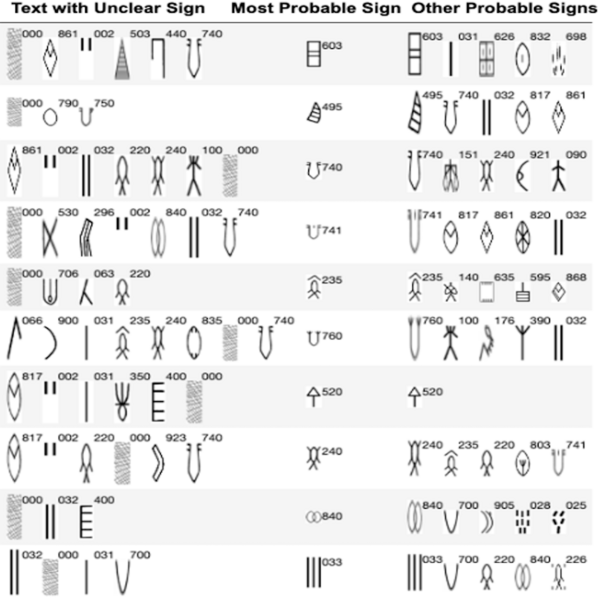
This study utilizes machine learning models to predict missing and unclear signs from the Indus script, a writing system from an ancient civilization in the Indian subcontinent.
Read More...Exponential regression analysis of the Canadian Zero Emission Vehicle market’s effects on climate emissions in 2030

Here, the authors explored how the sale and use of electric vehicles could reduce emissions from the transport industry in Canada. By fitting the sale of total of electric vehicles with an exponential model, the authors predicted the number of electric vehicle sales through 2030 and related that to the average emission for such vehicles. Ultimately, they found that the sale and use of electric vehicles alone would likely not meet the 45% reduction in emissions from the transport industry suggested by the Canadian government
Read More...Open Source RNN designed for text generation is capable of composing music similar to Baroque composers
Recurrent neural networks (RNNs) are useful for text generation since they can generate outputs in the context of previous ones. Baroque music and language are similar, as every word or note exists in context with others, and they both follow strict rules. The authors hypothesized that if we represent music in a text format, an RNN designed to generate language could train on it and create music structurally similar to Bach’s. They found that the music generated by our RNN shared a similar structure with Bach’s music in the input dataset, while Bachbot’s outputs are significantly different from this experiment’s outputs and thus are less similar to Bach’s repertoire compared to our algorithm.
Read More...Training neural networks on text data to model human emotional understanding

The authors train a neural network to detect text-based emotions including joy, sadness, anger, fear, love, and surprise.
Read More...The utilization of Artificial Intelligence in enabling the early detection of brain tumors

AI analysis of brain scans offers promise for helping doctors diagnose brain tumors. Haider and Drosis explore this field by developing machine learning models that classify brain scans as "cancer" or "non-cancer" diagnoses.
Read More...Temporal characterization of electroencephalogram slowing activity types
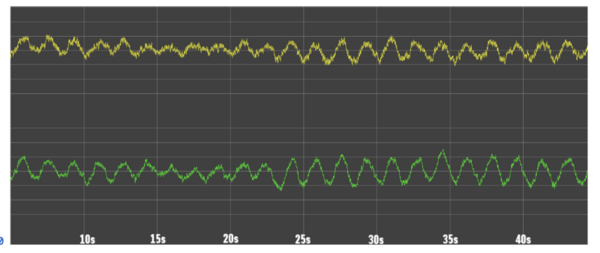
The authors use machine learning to analyze electroencephalogram data and identify slowing patterns that can indicate undetected disorders like epilepsy or dementia
Read More...