%20(1).png)
In this study, the authors identify transcripts and gene networks that are changed after infection with the Middle East Respiratory Syndrome-related coronavirus (MERS-CoV).
Read More...Gene expression profiling of MERS-CoV-London strain
%20(1).png)
In this study, the authors identify transcripts and gene networks that are changed after infection with the Middle East Respiratory Syndrome-related coronavirus (MERS-CoV).
Read More...Analysis of complement system gene expression and outcome across the subtypes of glioma
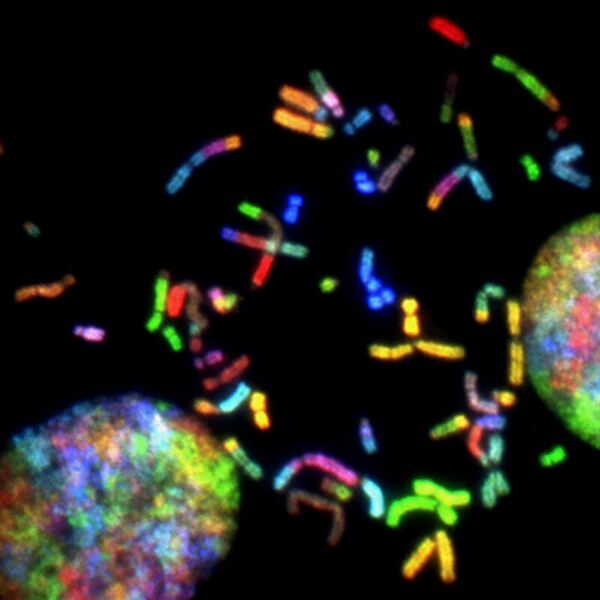
Here the authors sought to better understand glioma, cancer that occurs in the glial cells of the brain with gene expression profile analysis. They considered the expression of complement system genes across the transcriptional and IDH-mutational subtypes of low-grade glioma and glioblastoma. Based on their results of their differential gene expression analysis, they found that outcomes vary across different glioma subtypes, with evidence suggesting that categorization of the transcriptional subtypes could help inform treatment by providing an expectation for treatment responses.
Read More...Characterization of Inflammatory Cytokine Gene Expression in a Family with a History of Psoriasis
Psoriasis is a heritable autoimmune disorder characterized by abnormal red and itchy skin patches. The authors study the family of a man with psoriasis. They explore whether the man's children, who do not show any symptoms of psoriasis, demonstrate gene expression consistent with the disease.
Read More...Investigating KNOX Gene Expression in Aquilegia Petal Spur Development
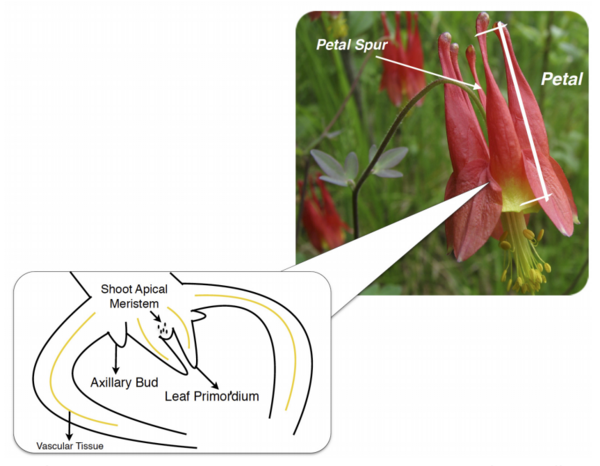
Plants, and all other multi-cellular organisms, develop through the coordinated action of many sets of genes. The authors here investigate the genes, in a class named KNOX, potentially responsible for organizing a certain part of Aquilegia (columbine) flowers called petal spurs. Through the technique Reverse Transcription-Polymerase Chain Reaction (RT-PCR), they find that certain KNOX genes are expressed non-uniformly in petal spurs, suggesting that they may be involved, perhaps in a cell-specific manner. This research will help guide future efforts toward understanding how many beautiful flowers develop their unique shapes.
Read More...Impact of TCERG1 SNP on gene expression and protein interactome in Huntington’s disease
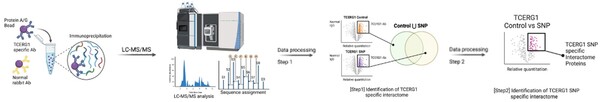
The authors assess a genetic variant within a well-known interaction partner of huntingtin that has been linked to modifying the age of onset of Huntington's disease.
Read More...The effects of the cancer metastasis promoting gene CD151 in E. coli
The independent effects of metastasis-promoting gene CD151 in the process of metastasis are not known. This study aimed to isolate CD151 to discover what its role in metastasis would be uninfluenced by potential interactions with other components and pathways in human cells. Results showed that CD151 significantly increased the adhesion of the cells and decreased their motility. Thus, it may be that CD151 is upregulated in cancer cells for the last step of metastasis, and it increases the chances of success of metastasis by aiding in implantation of the cancer cells. Targeting CD151 in chemotherapeutic modalities could therefore potentially slow or prevent metastasis.
Read More...The Effect of the Human MeCP2 gene on Drosophila melanogaster behavior and p53 inhibition as a model for Rett Syndrome

In this study, the authors observe if the symptoms of Rett Syndrome, a neurodegenerative disease in humans, are reflected in Drosophila melanogaster. This was achieved by differentiating the behavior and physical aspects of wild-type flies from flies expressing the full-length MeCP2 gene and the mutated MeCP2 gene (R106W). After conducting these experiments, some of the Rett Syndrome symptoms were recapitulated in Drosophila, and a subset of those were partially ameliorated by the introduction of pifithrin-alpha.
Read More...String analysis of exon 10 of the CFTR gene and the use of Bioinformatics in determination of the most accurate DNA indicator for CF prediction
Cystic fibrosis is a genetic disease caused by mutations in the CFTR gene. In this paper, the authors attempt to identify variations in stretches of up to 8 nucleotides in the protein-coding portions of the CFTR gene that are associated with disease development. This would allow screening of newborns or even fetuses in utero to determine the likelihood they develop cystic fibrosis.
Read More...Characterization and Phylogenetic Analysis of the Cytochrome B Gene (cytb) in Salvelinus fontinalis, Salmo trutta and Salvelinus fontinalis X Salmo trutta Within the Lake Champlain Basin

Recent declines in the brook trout population of the Lake Champlain Basin have made the genetic screening of this and other trout species of utmost importance. In this study, the authors collected and analyzed 21 DNA samples from Lake Champlain Basin trout populations and performed a phylogenetic analysis on these samples using the cytochrome b gene. The findings presented in this study may influence future habitat decisions in this region.
Read More...Investigating the effects of mutations of amino acids on the protein expression of CDK2 cancer gene