Impact of TCERG1 SNP on gene expression and protein interactome in Huntington’s disease
(1) Phillips Exeter Academy, (2) Molecular Neurogenetics Unit, Center for Genomic Medicine, Massachusetts General Hospital; Department of Neurology, Harvard Medical School
https://doi.org/10.59720/25-066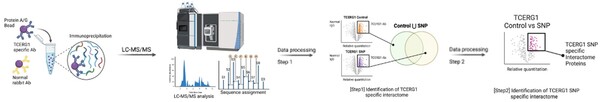
Huntington’s disease (HD) is a genetic neurodegenerative disorder that usually presents symptoms in adulthood. An expansion of the CAG repeat in the huntingtin gene (HTT) creates a mutated huntingtin protein, subsequently causing disease phenotypes. However, the disease age of onset is significantly influenced by single nucleotide polymorphisms (SNPs) across a patient’s genome. In this study, we investigated the role of the SNP rs79727797 located within TCERG1 (Transcription Elongation Regulator 1), a well-known huntingtin interactor, at the chromosome 5 locus 5BM1. We hypothesized that this SNP would influence gene expression, alter protein interactions, and affect cell growth. Using lymphoblastoid cell lines (LCLs) from HD patients with and without SNP rs79727797, we first assessed the effect of rs79727797 on TCERG1 and huntingtin protein levels and found no significant changes. Next, we optimized an immunoprecipitation protocol to isolate TCERG1 and any interacting proteins, allowing for a detailed analysis of potential SNP-driven changes in TCERG1 interactions. This study lays the foundation for future research studying how SNP rs79727797 modifies HD onset, including potential alterations to the TCERG1 interactome.
This article has been tagged with: