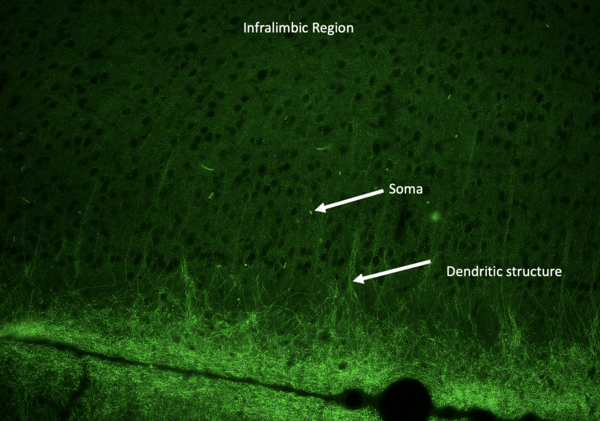
Although no comprehensive characterization of schizophrenia exists, there is a general consensus that patients have electrical dysfunction in the prefrontal cortex. The authors designed a novel piezoelectric silk-based implant and optimized electrical output through the addition of conductive materials zinc oxide (ZnO) and aluminum nitride (AlN). With further research and compatibility studies, this implant could rectify electrical misfiring in the infralimbic prefrontal cortex.
Read More...
.jpg)