The authors test different machine learning algorithms to remove background noise from audio to help people with hearing loss differentiate between important sounds and distracting noise.
Read More...Using neural networks to detect and categorize sounds

The authors test different machine learning algorithms to remove background noise from audio to help people with hearing loss differentiate between important sounds and distracting noise.
Read More...Quantitative analysis and development of alopecia areata classification frameworks
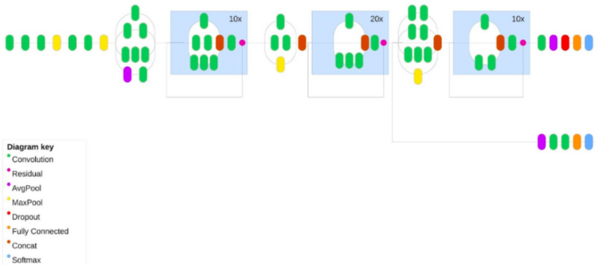
This article discusses Alopecia areata, an autoimmune disorder causing sudden hair loss due to the immune system mistakenly attacking hair follicles. The article introduces the use of deep learning (DL) techniques, particularly convolutional neural networks (CNN), for classifying images of healthy and alopecia-affected hair. The study presents a comparative analysis of newly optimized CNN models with existing ones, trained on datasets containing images of healthy and alopecia-affected hair. The Inception-Resnet-v2 model emerged as the most effective for classifying Alopecia Areata.
Read More...Prediction of diabetes using supervised classification
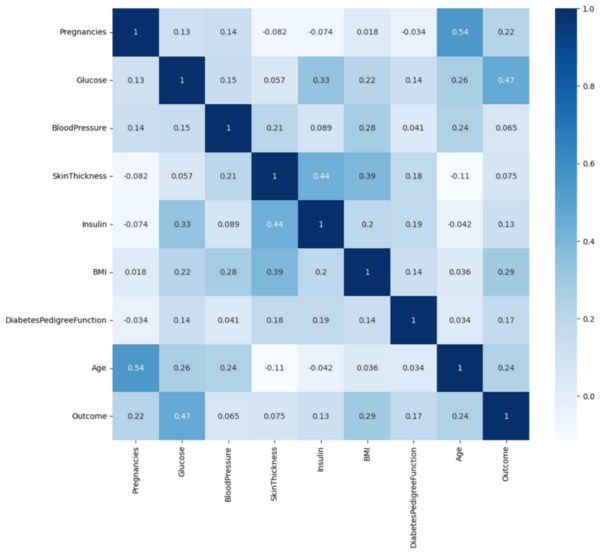
The authors develop and test a machine learning algorithm for predicting diabetes diagnoses.
Read More...Predicting baseball pitcher efficacy using physical pitch characteristics

Here, the authors sought to develop a new metric to evaluate the efficacy of baseball pitchers using machine learning models. They found that the frequency of balls, was the most predictive feature for their walks/hits allowed per inning (WHIP) metric. While their machine learning models did not identify a defining trait, such as high velocity, spin rate, or types of pitches, they found that consistently pitching within the strike zone resulted in significantly lower WHIPs.
Read More...Model selection and optimization for poverty prediction on household data from Cambodia

Here the authors sought to use three machine learning models to predict poverty levels in Cambodia based on available household data. They found teat multilayer perceptron outperformed the other models, with an accuracy of 87 %. They suggest that data-driven approaches such as these could be used more effectively target and alleviate poverty.
Read More...An explainable model for content moderation
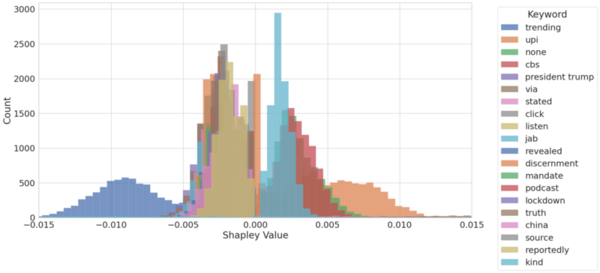
The authors looked at the ability of machine learning algorithms to interpret language given their increasing use in moderating content on social media. Using an explainable model they were able to achieve 81% accuracy in detecting fake vs. real news based on language of posts alone.
Read More...Evaluating the feasibility of SMILES-based autoencoders for drug discovery
The authors investigate the ability of machine learning models to developing new drug-like molecules by learning desired chemical properties versus simply generating molecules that similar to those in the training set.
Read More...Comparing model-centric and data-centric approaches to determine the efficiency of data-centric AI
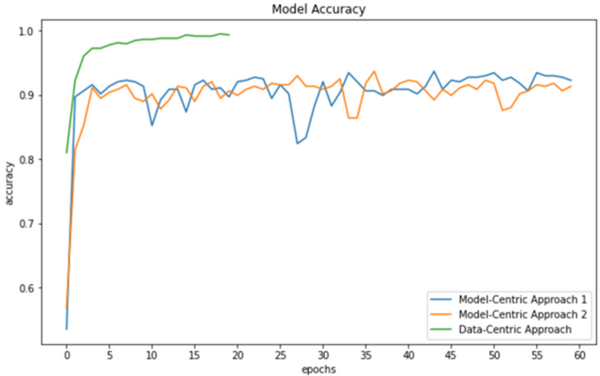
In this study, three models are used to test the hypothesis that data-centric artificial intelligence (AI) will improve the performance of machine learning.
Read More...Predicting the Instance of Breast Cancer within Patients using a Convolutional Neural Network
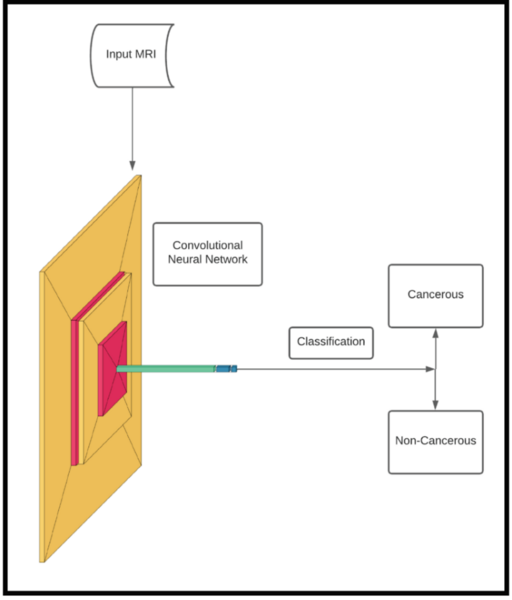
Using a convolution neural network, these authors show machine learning can clinically diagnose breast cancer with high accuracy.
Read More...The comparative effect of remote instruction on students and teachers

In this study, high school students and teachers responded to a survey consisting of Likert-type scale, multiple-choice, and open-ended questions regarding various aspects of remote instruction. After analyzing the data collected, they found that remote learning impacted high school students academically and socially. Students took longer to complete assignments, and both students and teachers felt that students do not learn as much in remote learning compared to in-person instruction. However, most high school students demonstrated a comprehensive understanding of the topics, and an overall negative impact on students' grades was not detected.
Read More...