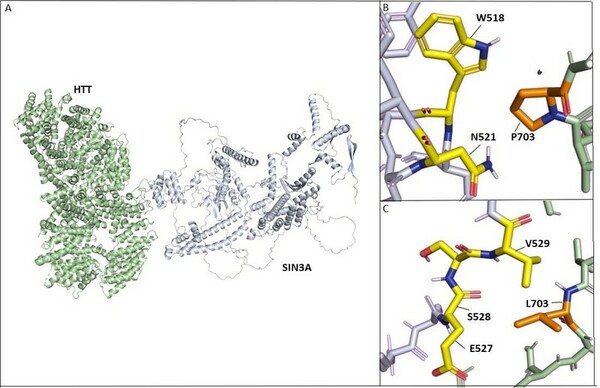
In an extensive study of gene mutations, and their resulting effect on protein-protein interactions, Desai and Stork found that HTT-PRPF40B-MECP2 interactions are weakened with progression of Lopes-Maciel-Rodan syndrome.
Read More...Disruptions in protein-protein interactions between HTT, PRPF40B, and MECP2 are involved in Lopes-Maciel-Rodan syndrome

In an extensive study of gene mutations, and their resulting effect on protein-protein interactions, Desai and Stork found that HTT-PRPF40B-MECP2 interactions are weakened with progression of Lopes-Maciel-Rodan syndrome.
Read More...Expressional correlations between SERPINA6 and pancreatic ductal adenocarcinoma-linked genes
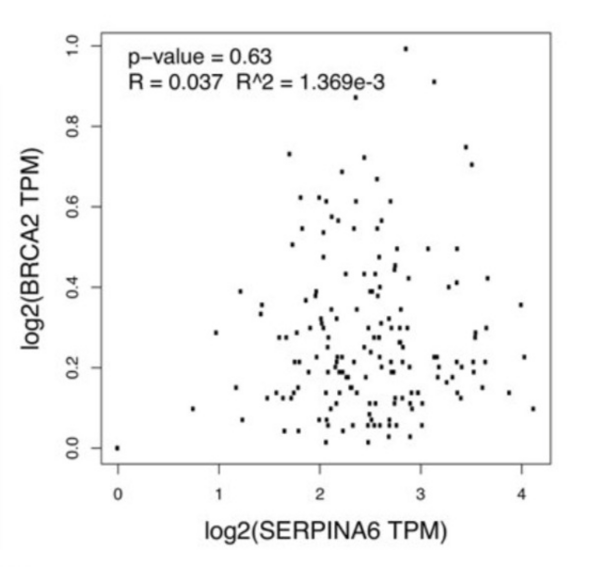
Pancreatic ductal adenocarcinoma (PDAC) is the most common form of pancreatic cancer, with early diagnosis and treatment challenges. When any of the genes KRAS, SMAD4, TP53, and BRCA2 are heavily mutated, they correlate with PDAC progression. Cellular stress, partly regulated by the gene SERPINA6, also correlates with PDAC progression. When SERPINA6 is highly expressed, corticosteroid-binding globulin inhibits the effect of the stress hormone cortisol. In this study, the authors explored whether there is an inverse correlation between the expression of SERPINA6 and PDAC-linked genes.
Read More...Combinatorial treatment by siNOTCH and retinoic acid decreases A172 brain cancer cell growth
Treatments inhibiting Notch signaling pathways have been explored by researchers as a new approach for the treatment of glioblastoma tumors, which is a fast-growing and aggressive brain tumor. Recently, retinoic acid (RA) therapy, which inhibits Notch signaling, has shown a promising effect on inhibiting glioblastoma progression. RA, which is a metabolite of vitamin A, is very important in embryonic cellular development, which includes the regulation of multiple developmental processes, such as brain neurogenesis. However, high doses of RA treatment caused many side effects such as headaches, nausea, redness around the injection site, or allergic reactions. Therefore, we hypothesized that a combination treatment of RA and siRNA targeting NOTCH1 (siNOTCH1), the essential gene that activates Notch signaling, would effectively inhibit brain cancer cell proliferation. The aim of the study was to determine whether inhibiting NOTCH1 would inhibit the growth of brain cancer cells by cell viability assay. We found that the combination treatment of siNOTCH1 and RA in low concentration effectively decreased the NOTCH1 expression level compared to the individual treatments. However, the combination treatment condition significantly decreased the number of live brain cancer cells only at a low concentration of RA. We anticipate that this novel combination treatment can provide a solution to the side effects of chemotherapy.
Read More...Quantifying natural recovery of dopamine deficits induced by chronic stress
Here the authors investigated the natural recovery of stress-induced dopamine-related gene deficits in C. elegans by measuring the expression of cat-2 (dopamine biosynthesis) and sod-2 (oxidative stress) following exposure to starvation or hydrocortisone. They found that the reversibility of sod-2 and the expression of cat-2 were highly dependent on the type and severity of the stressor, suggesting that the body's natural ability to recover from dopamine dysfunction has biological limitations.
Read More...Evolution of Neuroplastin-65
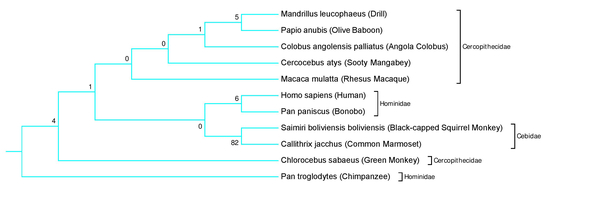
Human intelligence is correlated with variation in the protein neuroplastin-65, which is encoded by the NPTN gene. The authors examine the evolution of this gene across different animal species.
Read More...Upregulation of the Ribosomal Pathway as a Potential Blood-Based Genetic Biomarker for Comorbid Major Depressive Disorder (MDD) and PTSD
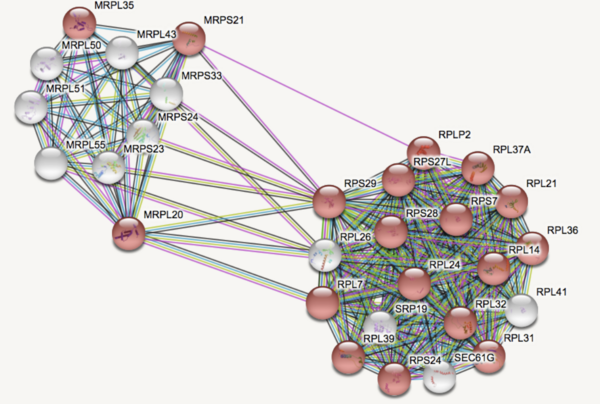
Major Depressive Disorder (MDD), and Post-Traumatic Stress Disorder (PTSD) are two of the fastest growing comorbid diseases in the world. Using publicly available datasets from the National Institute for Biotechnology Information (NCBI), Ravi and Lee conducted a differential gene expression analysis using 184 blood samples from either control individuals or individuals with comorbid MDD and PTSD. As a result, the authors identified 253 highly differentially-expressed genes, with enrichment for proteins in the gene ontology group 'Ribosomal Pathway'. These genes may be used as blood-based biomarkers for susceptibility to MDD or PTSD, and to tailor treatments within a personalized medicine regime.
Read More...TGFβ1 Codon 10 Polymorphism and its Association with the Prevalence of Low Myopia
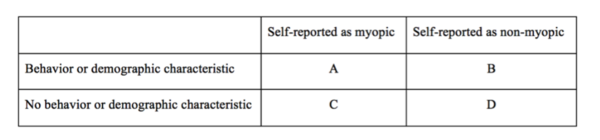
The goal of this project was to assess the relationships among low myopia, behavioral and demographic factors, and a single-nucleotide polymorphism (SNP) in the TGFβ1 gene.
Read More...Cutibacterium acnes sequence space topology implicates recA and guaA as potential virulence factors
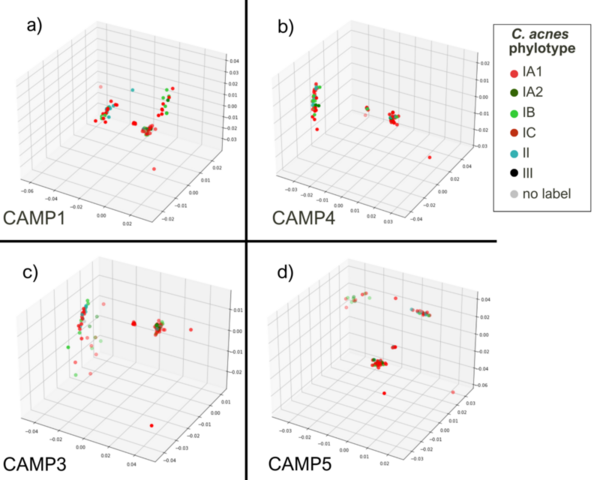
Cutibacterium acnes is a bacterium believed to play an important role in the pathogenesis of common skin diseases such as acne vulgaris. Currently, acne is known to be associated with strains from the type IA1 and IC clades of C. acnes, while those from the type IA2, IB, II, and III phylogroups are associated with skin health. This is the first study to explore the sequence space of individual gene products of different C. acnes phylogroups. Our analysis compared the sequence space topology of virulence factors to proteins with unknown functions and housekeeping proteins. We hypothesized that sequence space features of virulence factors are different from housekeeping protein features, which potentially provides an avenue to deduce unknown proteins’ functions. This proposition should be confirmed based on further experimental outcomes. A notable similarity in the sequence spaces’ topological features of previously known as housekeeping proteins encoded by recA and guaA genes to ‘putative virulence’ genes camp2 and tly was observed. Our research suggests further investigation of recA and guaA’s potential virulence properties to better understand acne pathogenesis and develop more targeted acne treatments.
Read More...Refinement of Single Nucleotide Polymorphisms of Atopic Dermatitis related Filaggrin through R packages

In the United States, there are currently 17.8 million affected by atopic dermatitis (AD), commonly known as eczema. It is characterized by itching and skin inflammation. AD patients are at higher risk for infections, depression, cancer, and suicide. Genetics, environment, and stress are some of the causes of the disease. With the rise of personalized medicine and the acceptance of gene-editing technologies, AD-related variations need to be identified for treatment. Genome-wide association studies (GWAS) have associated the Filaggrin (FLG) gene with AD but have not identified specific problematic single nucleotide polymorphisms (SNPs). This research aimed to refine known SNPs of FLG for gene editing technologies to establish a causal link between specific SNPs and the diseases and to target the polymorphisms. The research utilized R and its Bioconductor packages to refine data from the National Center for Biotechnology Information's (NCBI's) Variation Viewer. The algorithm filtered the dataset by coding regions and conserved domains. The algorithm also removed synonymous variations and treated non-synonymous, frameshift, and nonsense separately. The non-synonymous variations were refined and ordered by the BLOSUM62 substitution matrix. Overall, the analysis removed 96.65% of data, which was redundant or not the focus of the research and ordered the remaining relevant data by impact. The code for the project can also be repurposed as a tool for other diseases. The research can help solve GWAS's imprecise identification challenge. This research is the first step in providing the refined databases required for gene-editing treatment.
Read More...Genomic Signature Analysis for the Strategic Bioremediation of Polycyclic Aromatic Hydrocarbons in Mangrove Ecosystems in the Gulf of Tonkin

Engineered bacteria that degrade oil are currently being considered as a safe option for the treatment of oil spills. For this approach to be successful, the bacteria must effectively express oil-degrading genes they uptake as part of an external genoming vehicle called a "plasmid". Using a computational approach, the authors investigate plasmid-bacterium compatibility to find pairs that ensure high levels of gene expression.
Read More...