Upregulation of the Ribosomal Pathway as a Potential Blood-Based Genetic Biomarker for Comorbid Major Depressive Disorder (MDD) and PTSD
(1) International Academy, Bloomfield, Michigan, (2) miRcore, Ann Arbor, Michigan
https://doi.org/10.59720/18-005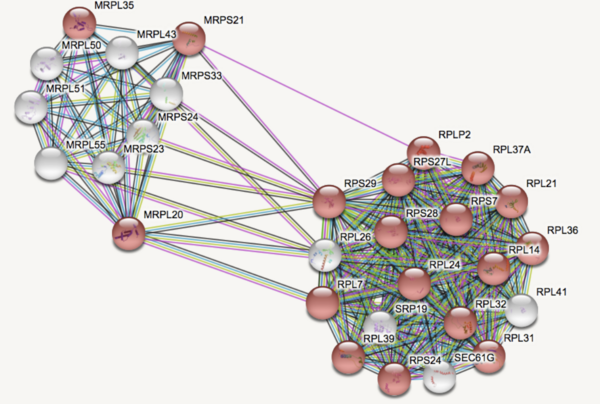
Major Depressive Disorder (MDD), and Post-Traumatic Stress Disorder (PTSD) are two of the fastest growing comorbid diseases in the world. A patient without any genetic predisposition to depression can live a healthy lifestyle but develop depression as a result of their environment, such as the occurrence of a traumatic event. This study addresses the question of whether there are any biomarkers in blood that could alert physicians of a patient’s susceptibility to comorbid MDD and PTSD. Since the pathophysiology of comorbid MDD is complex, it has been hypothesized that there could be several gene groups acting as biomarkers. Based on existing literature, the genes DICER1, FK506, and MRPS23 are thought to be relevant biomarkers. Using GEO2R, provided by NCBI, we conducted a differential gene expression analysis. We used patient dataset GSE67663, which contained 184 blood samples, including 72 control samples and 112 test samples. The top 253 genes that had p-values from 1.6 x 10^-6 to 3.49 x 10^3 were entered into the STRING database in order to identify common protein interactions. In accordance with the KEGG Ribosomal Pathway, two main gene groups, including those involved in mitochondrial ribosomal translation, were further analyzed for common gene ontology and biological processes. In the end, we found that the Ribosomal Pathway (ID: 03010) contained 29 upregulated genes and is a potential blood-based biomarker. This study has applications to the field of personalized medicine, in which tailored treatments based on biomarkers and one’s genome are utilized to identify people at risk for disease.
This article has been tagged with: