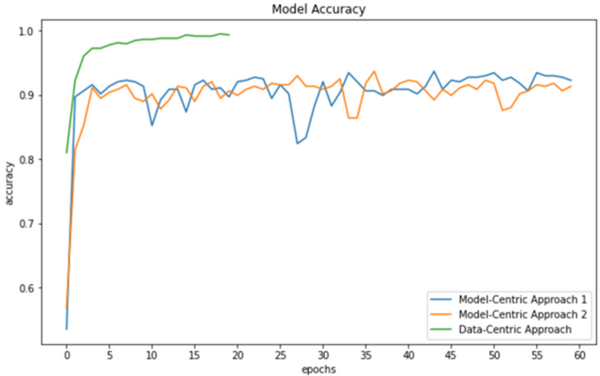
In this study, three models are used to test the hypothesis that data-centric artificial intelligence (AI) will improve the performance of machine learning.
Read More...Comparing model-centric and data-centric approaches to determine the efficiency of data-centric AI

In this study, three models are used to test the hypothesis that data-centric artificial intelligence (AI) will improve the performance of machine learning.
Read More...Similarity Graph-Based Semi-supervised Methods for Multiclass Data Classification
The purpose of the study was to determine whether graph-based machine learning techniques, which have increased prevalence in the last few years, can accurately classify data into one of many clusters, while requiring less labeled training data and parameter tuning as opposed to traditional machine learning algorithms. The results determined that the accuracy of graph-based and traditional classification algorithms depends directly upon the number of features of each dataset, the number of classes in each dataset, and the amount of labeled training data used.
Read More...Evaluating the effectiveness of synthetic training data for day-ahead wind speed prediction in the Great Lakes
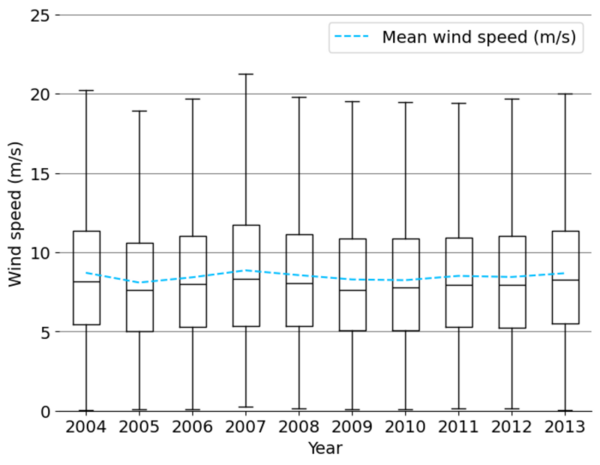
The authors looked at the feasibility to predict wind speeds that will have less reliance on using historical data.
Read More...Training neural networks on text data to model human emotional understanding

The authors train a neural network to detect text-based emotions including joy, sadness, anger, fear, love, and surprise.
Read More...Uncovering the hidden trafficking trade with geographic data and natural language processing

The authors use machine learning to develop an evidence-based detection tool for identifying human trafficking.
Read More...Predicting smoking status based on RNA sequencing data
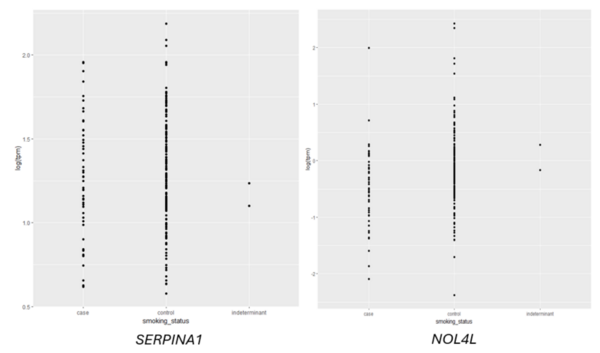
Given an association between nicotine addiction and gene expression, we hypothesized that expression of genes commonly associated with smoking status would have variable expression between smokers and non-smokers. To test whether gene expression varies between smokers and non-smokers, we analyzed two publicly-available datasets that profiled RNA gene expression from brain (nucleus accumbens) and lung tissue taken from patients identified as smokers or non-smokers. We discovered statistically significant differences in expression of dozens of genes between smokers and non-smokers. To test whether gene expression can be used to predict whether a patient is a smoker or non-smoker, we used gene expression as the training data for a logistic regression or random forest classification model. The random forest classifier trained on lung tissue data showed the most robust results, with area under curve (AUC) values consistently between 0.82 and 0.93. Both models trained on nucleus accumbens data had poorer performance, with AUC values consistently between 0.65 and 0.7 when using random forest. These results suggest gene expression can be used to predict smoking status using traditional machine learning models. Additionally, based on our random forest model, we proposed KCNJ3 and TXLNGY as two candidate markers of smoking status. These findings, coupled with other genes identified in this study, present promising avenues for advancing applications related to the genetic foundation of smoking-related characteristics.
Read More...Effects of different synthetic training data on real test data for semantic segmentation
Semantic segmentation - labelling each pixel in an image to a specific class- models require large amounts of manually labeled and collected data to train.
Read More...Heat conduction: Mathematical modeling and experimental data
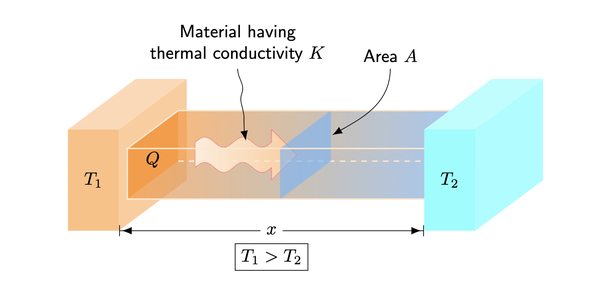
In this experiment, the authors modify the heat equation to account for imperfect insulation during heat transfer and compare it to experimental data to determine which is more accurate.
Read More...Machine learning on crowd-sourced data to highlight coral disease
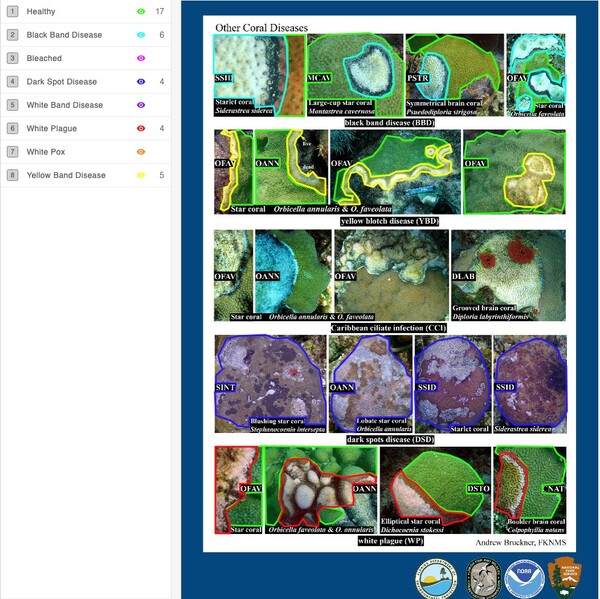
Triggered largely by the warming and pollution of oceans, corals are experiencing bleaching and a variety of diseases caused by the spread of bacteria, fungi, and viruses. Identification of bleached/diseased corals enables implementation of measures to halt or retard disease. Benthic cover analysis, a standard metric used in large databases to assess live coral cover, as a standalone measure of reef health is insufficient for identification of coral bleaching/disease. Proposed herein is a solution that couples machine learning with crowd-sourced data – images from government archives, citizen science projects, and personal images collected by tourists – to build a model capable of identifying healthy, bleached, and/or diseased coral.
Read More...LawCrypt: Secret Sharing for Attorney-Client Data in a Multi-Provider Cloud Architecture
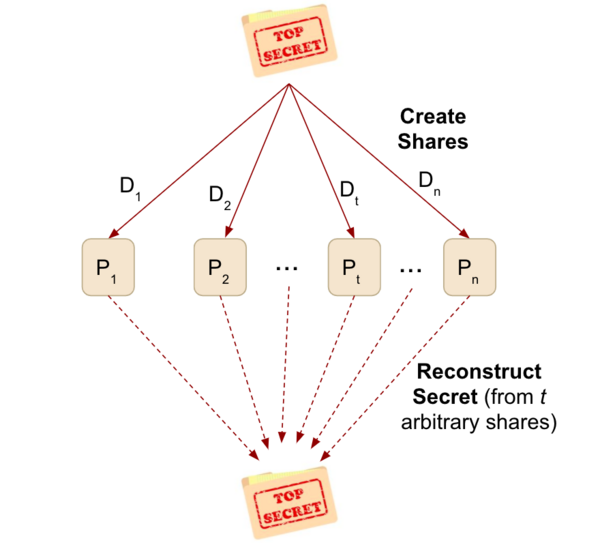
In this study, the authors develop an architecture to implement in a cloud-based database used by law firms to ensure confidentiality, availability, and integrity of attorney documents while maintaining greater efficiency than traditional encryption algorithms. They assessed whether the architecture satisfies necessary criteria and tested the overall file sizes the architecture could process. The authors found that their system was able to handle larger file sizes and fit engineering criteria. This study presents a valuable new tool that can be used to ensure law firms have adequate security as they shift to using cloud-based storage systems for their files.
Read More...