Differential MERS-CoV response in different cell types
(1) Saline High School, (2) Fulton Science Academy, (3) miRcore, (4) University of Michigan
https://doi.org/10.59720/22-096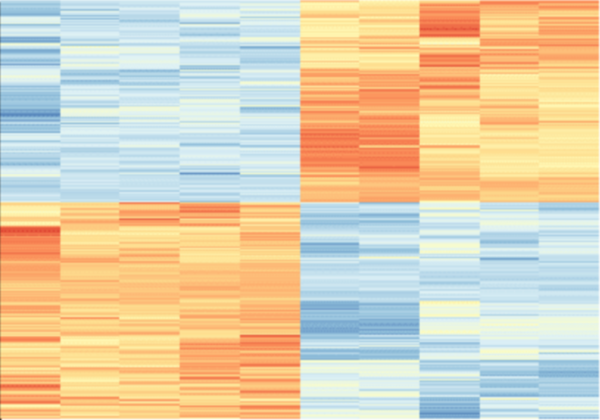
The ongoing COVID-19 pandemic piqued our interest in coronaviruses and prompted us to study MERS-CoV. While MERS-CoV has a low infection rate, it exhibits the highest mortality rate among the three known lethal coronaviruses. Thus, the potential for MERS-CoV to initiate a catastrophic pandemic remains a real threat. We decided to study MERS-CoV using online databases and analytical tools. We hypothesized that MERS-CoV infection elicits different responses in different types of host cells. We chose three data sets from Gene Omnibus Expression - GSE100504, GSE100509, and GSE86528, which represent MERS-CoV infection on human airway epithelial cells, primary human microvascular endothelial cells, and primary human fibroblasts, respectively. We performed RNA expression analysis on data collected 48 hours post-infection. Results indicated that human airway epithelial cells exhibited the most potent immune system response, suggesting that they are susceptible to pathogen infection, perhaps due to their location and exposure to the outside environment. We saw NRP1 and VEGFA upregulation in microvascular endothelial cells along with differential expression of angiogenesis and neural development processes. Given that NRP1 mediates SARS-CoV-2 entry into host cells, the infection mechanism for MERS-CoV on microvascular endothelial cells may be like that of SARS-CoV-2. In fibroblasts, we observed a positive regulation of the apoptotic process and negative regulation of cell proliferation. Together, our data supports our hypothesis and sheds light on the types of responses host cells elicit from MERS-CoV infection.
This article has been tagged with: