Analysis of antibiotic resistance genes in publicly accessible Staphylococcus aureus genomes
(1) Branham High School, (2) Cambridge University
https://doi.org/10.59720/23-333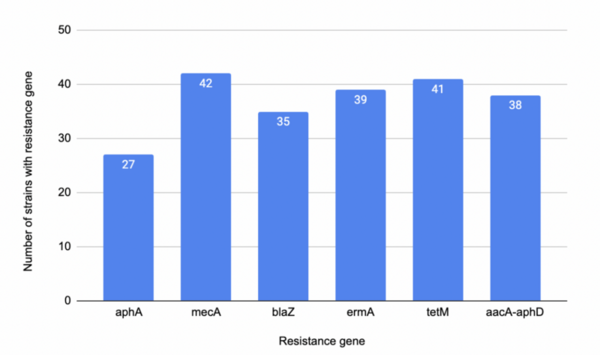
Staphylococcus aureus is a versatile bacterium commonly found in the human microbiota that can also cause a wide range of infections, from minor skin conditions to life-threatening diseases. Among its strains, methicillin-resistant Staphylococcus aureus (MRSA) is rapidly developing resistance to many antibiotics, including methicillin, penicillin, and other beta-lactam antibiotics. While MRSA incidence has declined in some areas, it remains a clinical threat due to its extensive resistance. Eradicating MRSA will take time, but a pressing question remains: can genetic diversity among MRSA strains guide the development of more effective treatments? We hypothesized that strains containing mecA and blaZ would show high antibiotic resistance, while strains with tetM would be least resistant. To test this, we used NCBI, PathogenWatch, and BLAST to identify and analyze 51 S. aureus strains and investigate their antibiotic resistance profiles. Our results showed that genetic diversity regarding resistance genes is present in all but six strains—those six lacked resistance genes entirely, making them highly susceptible to treatment. Our findings partially supported the hypothesis: mecA was strongly associated with resistance, tetM surprisingly also conferred resistance, and blaZ showed less resistance than expected. Our study underscores the need to examine genetic variability when designing treatments for MRSA. While no universal solution currently exists, understanding gene-based resistance patterns may eventually guide individualized treatment plans. Until then, combination antibiotic therapies may remain the most effective option against MRSA.
This article has been tagged with: