Uncovering mirror neurons’ molecular identity by single cell transcriptomics and microarray analysis
(1) YK Pao High School, Shanghai, China
https://doi.org/10.59720/22-080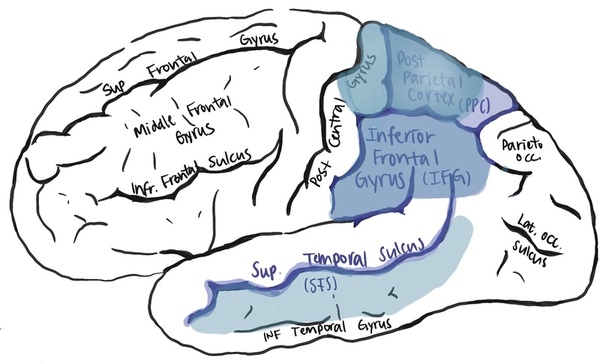
Mirror neurons (MNs) fire action potentials both when performing and seeing particular actions, ranging from grasping an object to observing a simple smile. During neuroimaging, the active cortical regions in both execution and observation are referred to as the mirror neuron system. It is composed of the superior temporal sulcus, posterior parietal cortex, and inferior frontal gyrus in humans. We aimed to find MNs’ neuronal identity, or the cell cluster to which MN belongs. Such identification lays the groundwork for future MN analysis and understanding of the mechanism of action. Due to the uniqueness of the human MN system, we performed bioinformatics analysis instead of experiments on animal models. High-expression genes throughout the MN system became candidate genes derived from differential gene analysis, or microarray. Ten cell clusters fit the MN’s trait as a layer three pyramidal excitatory neuron in the Allen Brain Atlas, human cortical single cell sequencing database. The cluster that had the highest sum of relative expression of candidate genes was cluster 85 with markers THEMIS and UBE2F. With MNs’ likely molecular identity in cluster 85 identified, we further explored MNs’ functional aspects. We gathered and analyzed data from single-cell sequencing and STRING (a functional protein association network) to uncover possible connections between typical neural degenerative diseases and psychiatric disorders with MN impairment. We found moebius facial syndrome, autism spectrum disorder, and amyotrophic lateral sclerosis to be potentially related to MN impairment.
This article has been tagged with: