Molecular Dynamics Simulations of Periplasmic Proteins Interacting With the Peptidoglycan Layer of Escherichia coli
(1) Lynbrook High School, San Jose, California, (2) The University of Iowa, Iowa City, Iowa
https://doi.org/10.59720/15-063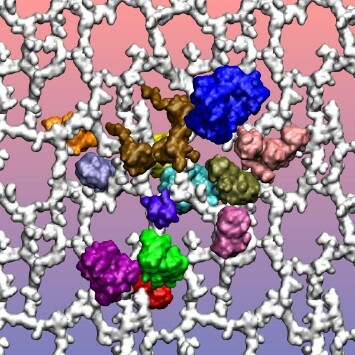
There has long been interest in using in silico methods to model complex biological systems, such as cells, for self-evident implications in molecular biology and medicine. Molecular dynamics (MD) simulations are ideal for the construction of such models due to the atomic resolution they provide. We use MD in this work to construct and simulate a preliminary model of the periplasmic space, the peptidoglycan layer and its associated proteins, in an E. coli cell. During an initial 23 nanoseconds (ns) MD simulation including 13 of the most abundant periplasmic proteins, the proteins and the peptidoglycan layer were characterized by several interactions. This work provides one of the first steps along the path to creating a complete and comprehensive model of a biological cell.
This article has been tagged with: