Design and in silico screening of analogs of rilpivirine as novel non-nucleoside reverse transcriptase inhibitors (NNRTIs) for antiretroviral therapy
(1) Mission San Jose High School, Fremont, CA, (2) Bishop O’Dowd High School, Oakland, CA, (3) Lynbrook High School, San Jose, CA, (4) Amador Valley High School, Pleasanton, CA, (5) American High School, Fremont, CA, (6) Aspiring Scholars Directed Research Program, Fremont, CA
https://doi.org/10.59720/20-218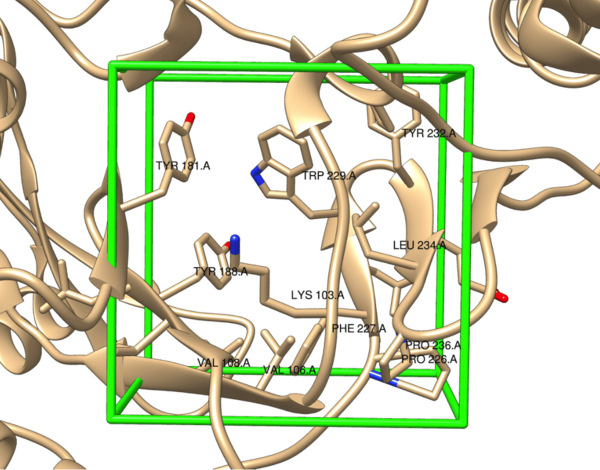
Part of the retroviral genus, the human immunodeficiency virus (HIV) relies on the host’s cellular machinery for replication. The viral genetic material is composed of ribonucleic acid (RNA) which, upon delivery into the host’s cells, is reverse transcribed into deoxyribonucleic acid (DNA) by the viral enzyme reverse transcriptase (RT). This is then used for the production of new viral components, thus allowing for further replication and spread of HIV. Because this virus selectively targets CD4+ T cells, the infection inevitably leads to weakening of the host’s immune system, which can then lead to acquired immunodeficiency syndrome (AIDS). One treatment for HIV is non-nucleoside reverse transcriptase inhibitors (NNRTIs), which are allosteric inhibitors of RT that disable the enzyme’s activity and thus viral replication. Previous NNRTIs approved for clinical use by the United States Food and Drug Association (FDA) include etravirine, doravirine, and rilpivirine. However, resistance to these drugs through mutations in RT necessitates the continued development of NNRTIs. Here, the structure of rilpivirine, a relatively recently FDA approved NNRTI, was used to design a library of analogs that were then evaluated in silico via high-throughput virtual screening (HTVS). From this, several structures were identified as potential next-generation NNRTIs with comparable predicted binding affinities to the allosteric binding pocket in RT as rilpivirine.
This article has been tagged with: