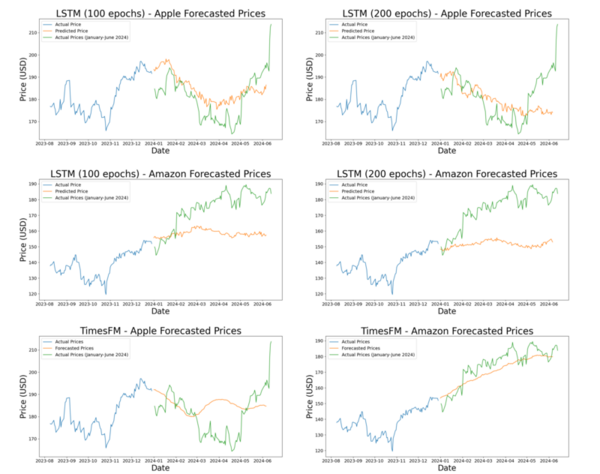
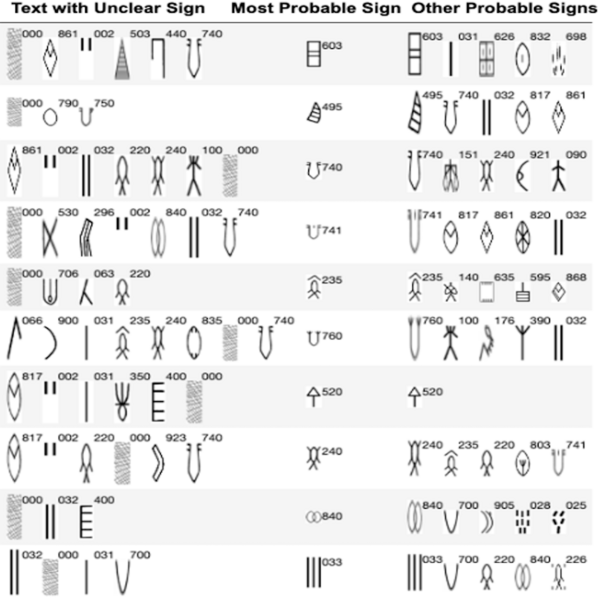
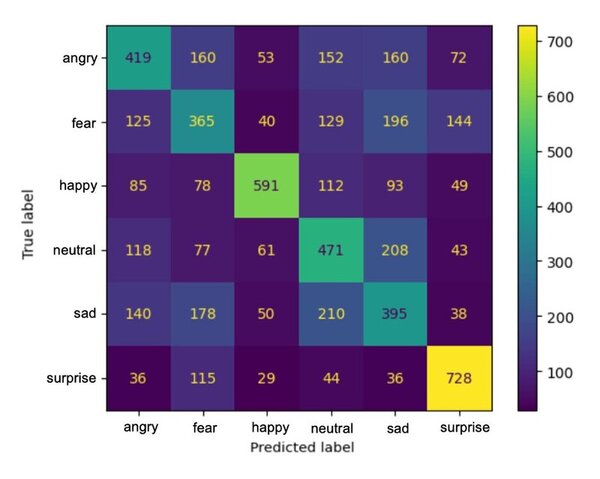
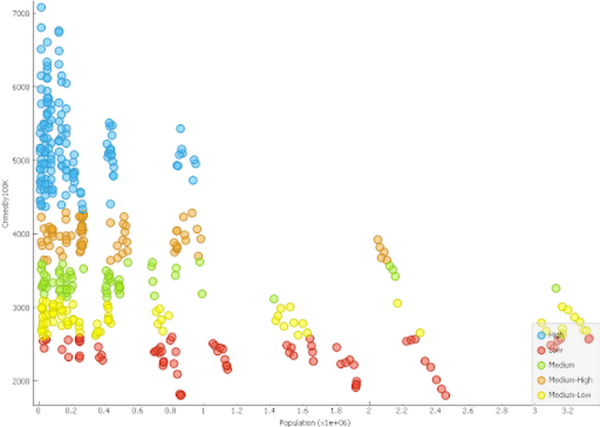
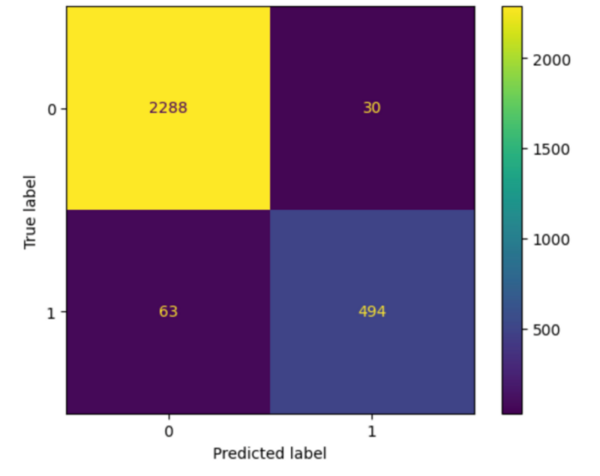
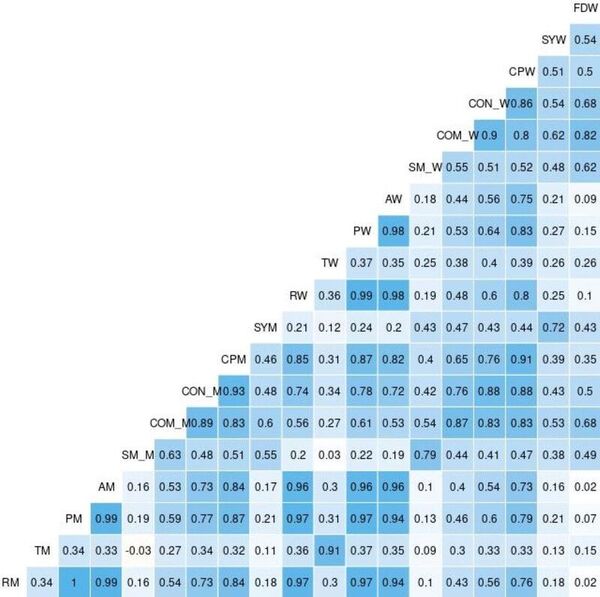
Here the authors sought to use three machine learning models to predict poverty levels in Cambodia based on available household data. They found teat multilayer perceptron outperformed the other models, with an accuracy of 87 %. They suggest that data-driven approaches such as these could be used more effectively target and alleviate poverty.
Read More...