
In this study, the effects of vitamin C, ginger, or curcumin supplements on C-reactive protein levels in healthy participants are determined in an eight-week open-label trial.
Read More...Reducing levels of C-Reactive Protein: An eight-week, open-label clinical trial of three oral supplements

In this study, the effects of vitamin C, ginger, or curcumin supplements on C-reactive protein levels in healthy participants are determined in an eight-week open-label trial.
Read More...Efficacy of Mass Spectrometry Versus 1H Nuclear Magnetic Resonance With Respect to Denaturant Dependent Hydrogen-Deuterium Exchange in Protein Studies
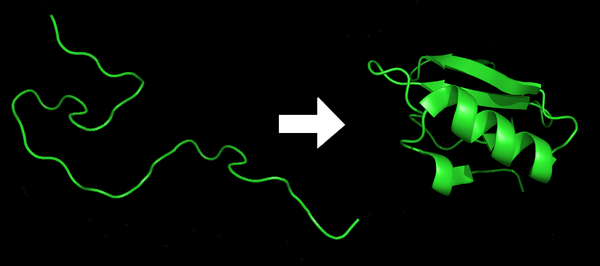
The misfolding of proteins leads to numerous diseases including Akzheimer’s, Parkinson’s and Type II Diabetes. Understanding of exactly how proteins fold is crucial for many medical advancements. Chenna and Englander addressed this problem by measuring the rate of hydrogen-deuterium exchange within proteins exposed to deuterium oxide in order to further elucidate the process of protein folding. Here, mass spectrometry was used to measure exchange in Cytochrome c and was compared to archived 1H NMR data.
Read More...Fluorescein or Green Fluorescent Protein: Is It Possible to Create a Sensor for Dehydration?

Currently there is no early dehydration detection system using temperature and pH as indicators. A sensor could alert the wearer and others of low hydration levels, which would normally be difficult to catch prior to more serious complications resulting from dehydration. In this study, a protein fluorophore, green fluorescent protein (GFP), and a chemical fluorophore, fluorescein, were tested for a change in fluorescence in response to increased temperature or decreased pH. Reversing the pH change did not restore GFP fluorescence, but that of fluorescein was re-established. This finding suggests that fluorescein could be used as a reusable sensor for a dehydration-related pH change.
Read More...Impact of TCERG1 SNP on gene expression and protein interactome in Huntington’s disease
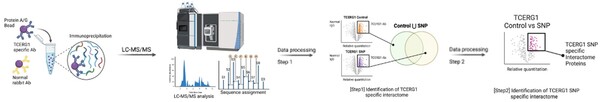
The authors assess a genetic variant within a well-known interaction partner of huntingtin that has been linked to modifying the age of onset of Huntington's disease.
Read More...The Cilium- and Centrosome-Associated Protein CCDC11 Is Required for Cytokinesis via Midbody Recruitment of the ESCRT- III Membrane Scission Complex Subunit CHMP2A
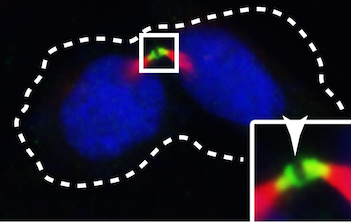
In order for cells to successfully multiply, a number of proteins are needed to correctly coordinate the replication and division process. In this study, students use fluorescence microscopy and molecular methods to study CCDC11, a protein critical in the formation of cilia. Interestingly, they uncover a new role for CCDC11, critical in the cell division across multiple human cell lines.
Read More...Investigating the effects of mutations of amino acids on the protein expression of CDK2 cancer gene
Mutation of the Catalytic Cysteine in Anopheles gambiae Transglutaminase 3 (AgTG3) Abolishes Plugin Crosslinking Activity without Disrupting Protein Folding Properties
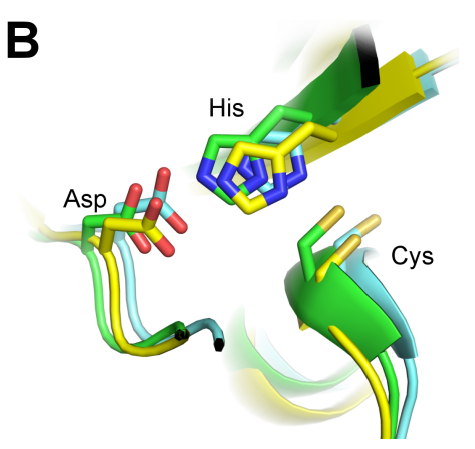
Malaria is a major public health issue, especially in developing countries, and vector control is a major facet of malaria eradication efforts. Recently, sterile insect technique (SIT), or the release of sterile mosquitoes into the wild, has shown significant promise as a method of keeping vector populations under control. In this study, the authors investigate the Anopheles gambiae transglutaminase 3 protein (AgT3), which is essential to the mating of the Anopheles mosquito. They show that an active site mutation is able to abolish the activity of the AgT3 enzyme and propose it as a potential target for chemosterilant inhibitors.
Read More...Evolution of Neuroplastin-65
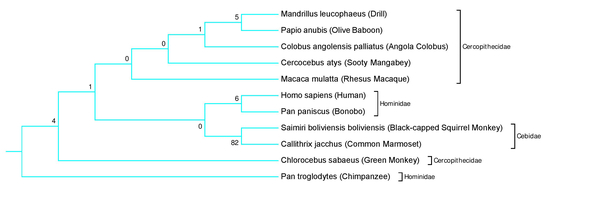
Human intelligence is correlated with variation in the protein neuroplastin-65, which is encoded by the NPTN gene. The authors examine the evolution of this gene across different animal species.
Read More...SOS-PVCase: A machine learning optimized lignin peroxidase with polyvinyl chloride (PVC) degrading properties
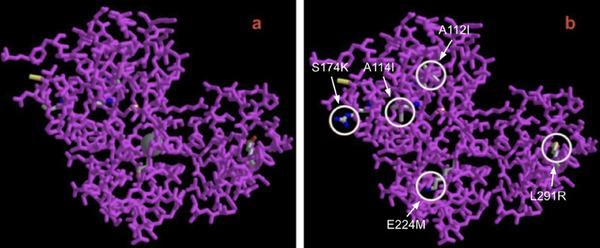
The authors looked at the primary structure of lignin peroxidase in an attempt to identify mutations that would improve both the stability and solubility of the peroxidase protein. The goal is to engineer peroxidase enzymes that are stable to help break down polymers, such as PVC, into monomers that can be reused instead of going to landfills.
Read More...The novel function of PMS2 mutation on ovarian cancer proliferation
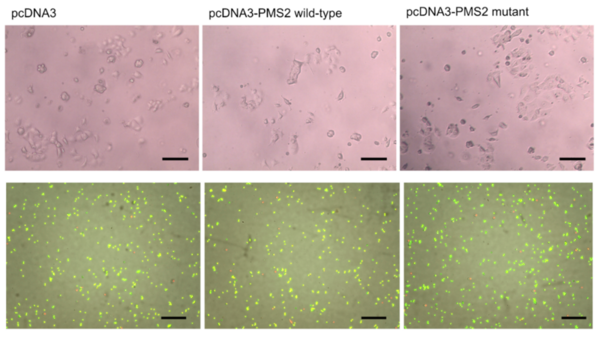
With disruption of DNA repair pathways pertinent to the timeline of cancer, thorough evaluation of mutations relevant to DNA repair proteins is crucial within cancer research. One such mutation includes S815L PMS2 - a mutation that results in significant decrease of DNA repair function by PMS2 protein. While mutation of PMS2 is associated with significantly increased colorectal and endometrial cancer risk, much work is left to do to establish the functional effects of the S815L PMS2 mutation in ovarian cancer progression. In this article, researchers contribute to this essential area of research by uncovering the tumor-progressive effects of the S815L PMS2 mutation in the context of ovarian cancer cell lines.
Read More...