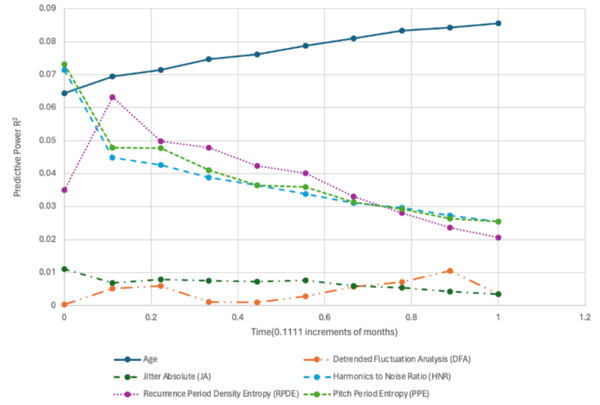
The authors looked at the ability to use audio clips to analyze the progression of Parkinson's disease.
Read More...Using advanced machine learning and voice analysis features for Parkinson’s disease progression prediction

The authors looked at the ability to use audio clips to analyze the progression of Parkinson's disease.
Read More...Exploring the effects of diverse historical stock price data on the accuracy of stock price prediction models
Algorithmic trading has been increasingly used by Americans. In this work, we tested whether including the opening, closing, and highest prices in three supervised learning models affected their performance. Indeed, we found that including all three prices decreased the error of the prediction significantly.
Read More...String analysis of exon 10 of the CFTR gene and the use of Bioinformatics in determination of the most accurate DNA indicator for CF prediction
Cystic fibrosis is a genetic disease caused by mutations in the CFTR gene. In this paper, the authors attempt to identify variations in stretches of up to 8 nucleotides in the protein-coding portions of the CFTR gene that are associated with disease development. This would allow screening of newborns or even fetuses in utero to determine the likelihood they develop cystic fibrosis.
Read More...Validating DTAPs with large language models: A novel approach to drug repurposing

Here, the authors investigated the integration of large language models (LLMs) with drug target affinity predictors (DTAPs) to improve drug repurposing, demonstrating a significant increase in prediction accuracy, particularly with GPT-4, for psychotropic drugs and the sigma-1 receptor. This novel approach offers to potentially accelerate and reduce the cost of drug discovery by efficiently identifying new therapeutic uses for existing drugs.
Read More...Monitoring drought using explainable statistical machine learning models
Droughts have a wide range of effects, from ecosystems failing and crops dying, to increased illness and decreased water quality. Drought prediction is important because it can help communities, businesses, and governments plan and prepare for these detrimental effects. This study predicts drought conditions by using predictable weather patterns in machine learning models.
Read More...Using broad health-related survey questions to predict the presence of coronary heart disease
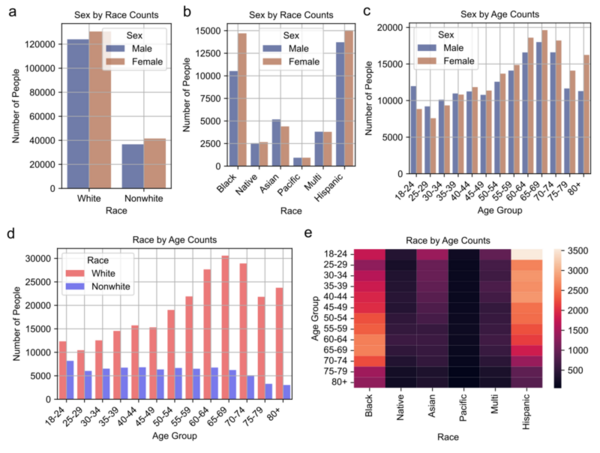
Coronary heart disease (CHD) is the leading cause of death in the U.S., responsible for nearly 700,000 deaths in 2021, and is marked by artery clogging that can lead to heart attacks. Traditional prediction methods require expensive clinical tests, but a new study explores using machine learning on demographic, clinical, and behavioral survey data to predict CHD.
Read More...Contrasting role of ASCC3 and ALKBH3 in determining genomic alterations in Glioblastoma Multiforme
Glioblastoma Multiforme (GBM) is the most malignant brain tumor with the highest fraction of genome alterations (FGA), manifesting poor disease-free status (DFS) and overall survival (OS). We explored The Cancer Genome Atlas (TCGA) and cBioportal public dataset- Firehose legacy GBM to study DNA repair genes Activating Signal Cointegrator 1 Complex Subunit 3 (ASCC3) and Alpha-Ketoglutarate-Dependent Dioxygenase AlkB Homolog 3 (ALKBH3). To test our hypothesis that these genes have correlations with FGA and can better determine prognosis and survival, we sorted the dataset to arrive at 254 patients. Analyzing using RStudio, both ASCC3 and ALKBH3 demonstrated hypomethylation in 82.3% and 61.8% of patients, respectively. Interestingly, low mRNA expression was observed in both these genes. We further conducted correlation tests between both methylation and mRNA expression of these genes with FGA. ASCC3 was found to be negatively correlated, while ALKBH3 was found to be positively correlated, potentially indicating contrasting dysregulation of these two genes. Prognostic analysis showed the following: ASCC3 hypomethylation is significant with DFS and high ASCC3 mRNA expression to be significant with OS, demonstrating ASCC3’s potential as disease prediction marker.
Read More...Determining degree of dissociation through conductivity
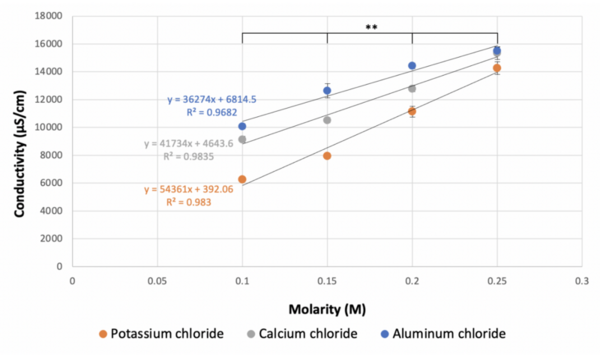
The authors looked at how molarity impacts the degree to which ionic compounds dissociate in solution. They found that lower molarities led to decreased conductivity of solutions in a manner that did not follow the theoretical predictions.
Read More...Using explainable artificial intelligence to identify patient-specific breast cancer subtypes
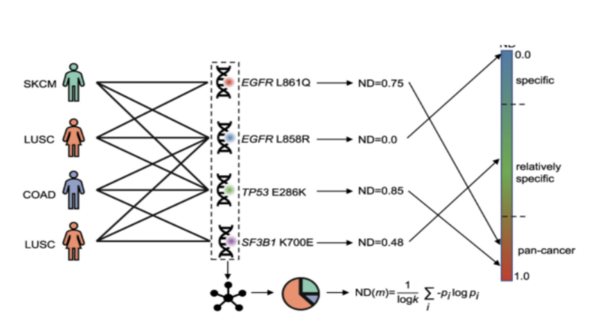
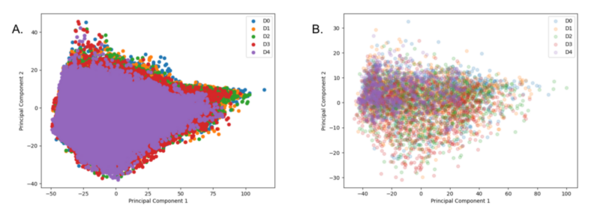
Breast cancer is the most common cancer in women, with approximately 300,000 diagnosed with breast cancer in 2023. It ranks second in cancer-related deaths for women, after lung cancer with nearly 50,000 deaths. Scientists have identified important genetic mutations in genes like BRCA1 and BRCA2 that lead to the development of breast cancer, but previous studies were limited as they focused on specific populations. To overcome limitations, diverse populations and powerful statistical methods like genome-wide association studies and whole-genome sequencing are needed. Explainable artificial intelligence (XAI) can be used in oncology and breast cancer research to overcome these limitations of specificity as it can analyze datasets of diagnosed patients by providing interpretable explanations for identified patterns and predictions. This project aims to achieve technological and medicinal goals by using advanced algorithms to identify breast cancer subtypes for faster diagnoses. Multiple methods were utilized to develop an efficient algorithm. We hypothesized that an XAI approach would be best as it can assign scores to genes, specifically with a 90% success rate. To test that, we ran multiple trials utilizing XAI methods through the identification of class-specific and patient-specific key genes. We found that the study demonstrated a pipeline that combines multiple XAI techniques to identify potential biomarker genes for breast cancer with a 95% success rate.
Read More...Increasing Average Yearly Temperature in Two U.S. Cities Shows Evidence for Climate Change
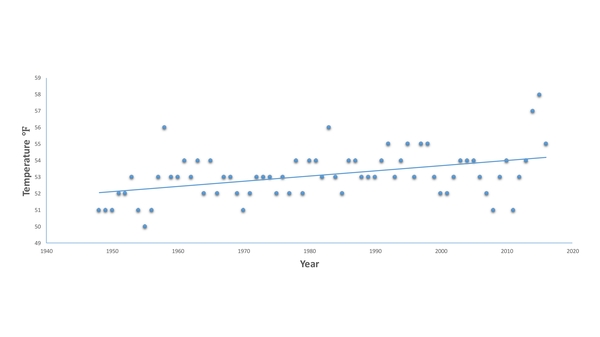
The authors were interested in whether they could observe the effects of climate change by analyzing historical temperature data of two U.S. cities. They predicted that they should observe a warming trend in both cities. Their results showed that despite yearly variations, warming trends can be observed both in Rochester, NY and Seattle, WA which fit the predictions of climate change forecasts.
Read More...