The impact of genetic analysis on the early detection of colorectal cancer
(1) Whitney M. Young Magnet High School, Chicago, Illinois, (2) University of Chicago, Biological Sciences Collegiate Division, Chicago, Illinois
https://doi.org/10.59720/22-174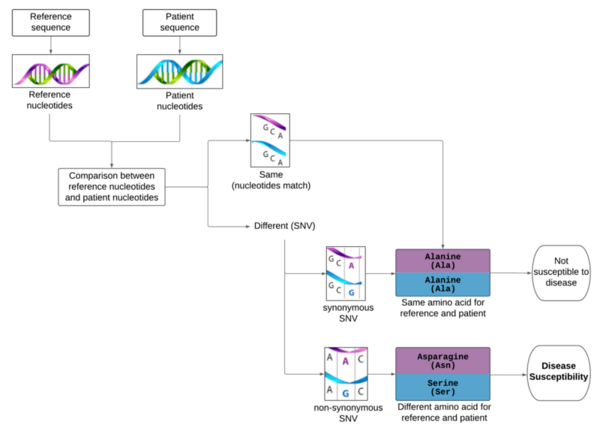
Although the 5-year survival rate for colorectal cancer is below 10%, it increases to greater than 90% if it is diagnosed early. We hypothesized from our research that analyzing non-synonymous single nucleotide variants (SNVs) in a patient's exome sequence would be an indicator for high genetic risk of developing colorectal cancer. First, the patient's exome sequence and the reference exome sequence were repeatedly aligned to identify the regions of similarity. The alignment between the two sequences that resulted in the most similar regions (optimal alignment) was selected. Next, we performed variant calling and identified variants in the patient's exome sequence. We applied a quality control check to assess sequencing data quality and filtered out the variants that did not pass the quality control check. Finally, the remaining selected variants were annotated with biologically pertinent information and explored for their potential roles in human disease by cross-referencing databases. A variant in the FGFR4 gene, known to cause accelerated cancer progression and tumor cell motility, was found in the patient's exome sequence. Studies suggest that this variant corresponds with an increased risk of colorectal cancer, supporting the usefulness of this procedure in early detection of colorectal cancer. This research was expanded to demonstrate that exome sequencing methods are capable of identifying other genetic variants. With higher computational power, one can produce more accurate alignment readings, detecting even the smallest of deviations from the reference exome sequence and thus enhancing the ability to evaluate genetic risk of disease.
This article has been tagged with: