Characterizing the evolution of antibiotic resistance in commercial Lactobacillus strains
(1) Olentangy High School, Lewis Center, Ohio
https://doi.org/10.59720/21-035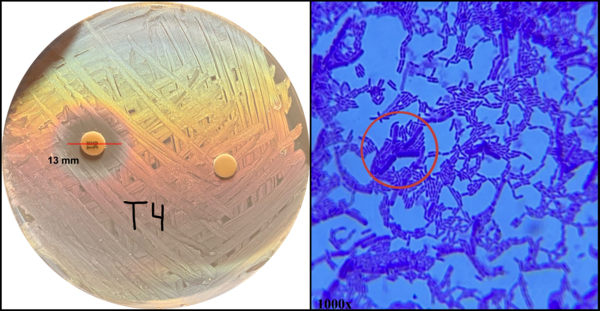
Throughout the world, millions of patients consume probiotic supplements and fermented foods for their perceived health benefits. Recent research suggests that the ingestion of bacteria commonly associated with probiotics negatively correlates with the prevalence of health conditions such as antibiotic-associated dysbiosis and can benefit immunocompromised patients through competitive inhibition. Therefore, healthcare providers often recommend co-administration of probiotics and antibiotics during treatment. Despite the benefits conferred by co-administration, the transfer of antibiotic-resistance genes from probiotics to pathogenic bacteria residing in the gut microbiome remains a significant risk to patient health, as many species are known to be resistant. We sought to determine how antibiotic resistance and its transferability developed among a commonly consumed species of probiotic, Lactobacillus. We hypothesized that these bacteria would demonstrate increased resistance to an antibiotic when administered over successive generations, that resistance would specifically develop during the first two generations of antibiotic exposure, and that antibiotic resistance could be modeled to predict future behavior in response to other antibiotics. We determined the transferability of antibiotic resistance by exposing Escherichia coli to resistant Lactobacillus and testing for antibiotic resistance. Our results demonstrated that Lactobacillus develops antibiotic resistance logarithmically with each generation and variably depending on the class of the antibiotic administered. Further, we created a model to predict how antibiotic resistance developed. Our findings may influence how healthcare providers evaluate the risks associated with probiotic co-administration in immunocompromised patients.
This article has been tagged with: